













| General Information: |
|
| Name(s) found: |
MMS22 /
YLR320W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1454 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
response to drug
[IMP double-strand break repair [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDVDEPNPIV ISDSEATDEE ISIIYEPEFN ENYLWAEENV QEASRSQKIV TERLSLDSTA 60
61 GESCTPSVVT DTQVTTGLRW SLRKRKAIQK MPYSLERIKH RQLLEGYDIS SFDSISNQLT 120
121 LPKNASTVIH SNDILLTKRT GKPLDEQKDV TIDSIKPENS SVQSQRYDSD EEIPKKRHRT 180
181 FKDLDQDIVF QSGDSTEDEQ DLASTNLQNT QNDEVIFRGR VLNVRTGYRG VLPRVAWEKS 240
241 LQKQQSSKVT KRKTQLLNHK GVAKRKMNRS AHIEDEEQNL LNDLIAPDDE LDIEENAPPD 300
301 IYLGNLPEDR EANEKELKEL QEYYESKYSE DAQSAGTSGF NLNEEYRNEP VYELEYDGPG 360
361 SCISHVSYKD QPIIYLNSRH SDSGFSEQYN ISAEDNQSVI SLDAAEEHND GIIDKMLVKP 420
421 KRIKATNDAN FLNTKSKRVR RYKYKYRNSC LAPSTKAIKV GKRSAHKSHL AANNPVSFVS 480
481 KKNHVIDDYF FEELESQSLE QDDSSSLKPQ KKRRKKKAPI YSSFSADLES RRKPVFNTVV 540
541 EVPTNRYAFT KPNVRNRDSI NHDMEFEEED SNQELGPIMV VLDSILLKKP FEPPNFFKIQ 600
601 LSDKSFLLSK LNPADIATSL QKIFRVIIDK GITDTELVHF NESLIAFLVH LDMPELFDLI 660
661 GEFHREFRSK VNSLRKKAKP IHFFQIAACQ LMFLEISRYN KISAAAKFDM DVKLLDHIVS 720
721 FFKLLSVCYD SVMKNPMQYL YTSYYILSAV VDVIHKKEAL WDLFQKHPFS PHISLLLVNI 780
781 FPTKVCRWQV LRLDSEFQPL SSAFRFINYC IETCNWNVTN SLILSLDRIF KRRRFSDFEE 840
841 ESDLSQNNKI IYPPTNQLTS RLMFNRYLHL LTLCELSSSD TQRVIPMGDI SMNDSLSVLK 900
901 NRLNLLIVLA TRFDLNLEKR FQELTRPLYS KEYLNLHTQN TVRTITTLIM QASLSFLEIS 960
961 RIKNHPFSGK FIASLFDKLV LQQPSISGVT ENFLKEFTNL VSKMKRKSVS MLKFLYPSLV1020
1021 AMSQENIFES SFFLLLQVYL KSLDVLGPTW VQNYLFQFIK SKAQENERWI ECYCQIGKFL1080
1081 VDSGIFTWWT FFTYNGLDAA LHFQLAFHSL IIDFCDTDSF ELLKKPLYSI ASDLLLISKD1140
1141 DAFYHFLSNL LKRAHIIVAD LKPVSDENEL LRLAYIFSKA LKKNAYQDLL AVFLSLAKKH1200
1201 YDEGDISRNF LAKYLEFLNK NCLTELRNNQ LFISLRRELG ISSDEDEKCA FWDSFNEAGD1260
1261 ILSKAAFVET GIVQACCTGN EIDGYLDNLS TLFTSTMLES PFAFFSDLVI AHIFENRPFF1320
1321 DVNIKNFLLS HFIDLFNKVL KMKFEQVSPD EFAELCKVYR ALCIECATDD TFNSNSDLIA1380
1381 AKDAFLVSVL RIADGFWEHD KLLQLRMLDS NMNIPNQIPH TTLQSSLSAI VIKIIESNIG1440
1441 KIEASEPFKT FKNT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
MMS22 YMR187C |
| View Details | Ho Y, et al. (2002) |
|
GDH2 MMS22 RTT101 RTT107 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
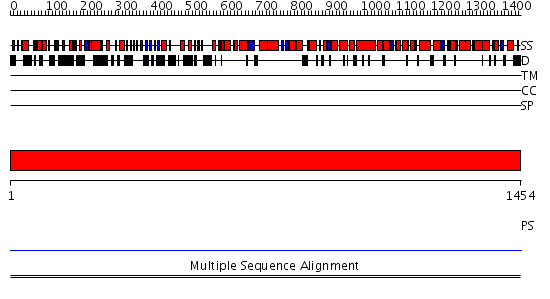
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1454] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [296..518] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [519..700] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [701..847] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [848..938] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [939..1165] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1166..1454] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)