













| General Information: |
|
| Name(s) found: |
FHLA_ECOLI
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli |
| Length: | 692 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYTPMSDLG QQGLFDITRT LLQQPDLASL CEALSQLVKR SALADNAAIV LWQAQTQRAS 60
61 YYASREKDTP IKYEDETVLA HGPVRSILSR PDTLHCSYEE FCETWPQLDA GGLYPKFGHY 120
121 CLMPLAAEGH IFGGCEFIRY DDRPWSEKEF NRLQTFTQIV SVVTEQIQSR VVNNVDYELL 180
181 CRERDNFRIL VAITNAVLSR LDMDELVSEV AKEIHYYFDI DDISIVLRSH RKNKLNIYST 240
241 HYLDKQHPAH EQSEVDEAGT LTERVFKSKE MLLINLHERD DLAPYERMLF DTWGNQIQTL 300
301 CLLPLMSGDT MLGVLKLAQC EEKVFTTTNL NLLRQIAERV AIAVDNALAY QEIHRLKERL 360
361 VDENLALTEQ LNNVDSEFGE IIGRSEAMYS VLKQVEMVAQ SDSTVLILGE TGTGKELIAR 420
421 AIHNLSGRNN RRMVKMNCAA MPAGLLESDL FGHERGAFTG ASAQRIGRFE LADKSSLFLD 480
481 EVGDMPLELQ PKLLRVLQEQ EFERLGSNKI IQTDVRLIAA TNRDLKKMVA DREFRSDLYY 540
541 RLNVFPIHLP PLRERPEDIP LLAKAFTFKI ARRLGRNIDS IPAETLRTLS NMEWPGNVRE 600
601 LENVIERAVL LTRGNVLQLS LPDIVLPEPE TPPAATVVAL EGEDEYQLIV RVLKETNGVV 660
661 AGPKGAAQRL GLKRTTLLSR MKRLGIDKSA LI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
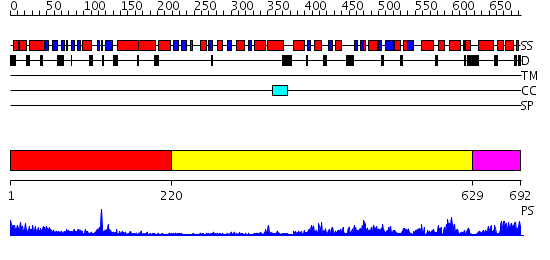
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..219] | 21.30103 | 3',5'-cyclic nucleotide phosphodiesterase 2A, GAF A and GAF B domains | |
| 2 | View Details | [220..628] | 91.39794 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 3 | View Details | [629..692] | 89.69897 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)