













| General Information: |
|
| Name(s) found: |
nas-30 /
CE36770
[WormBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 710 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA |
| Biological Process: |
proteolysis
[IEA |
| Molecular Function: |
meprin A activity
[IEA zinc ion binding [IEA astacin activity [IEA metallopeptidase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYMIQGCYIP AKSDKILETI FSDAPPVYRQ PRTKAEKIER FRNIARTFSP FVYEVNTTPA 60
61 PHFDNFIWQQ NAPAVTPEPF TFAPFSFPTL ATVAPPAPGP GGPTLEPFLP TTASPKLLAH 120
121 NTARMIREIA SFSDGGRSRD QDFGAVQTLM QAFFEAVSSG NNGGAGAAAG AGTALGDAPM 180
181 LQAHRDGTEL GANRALTNKL FESDMVLTVK QMKAIVLAAQ EARNPHGRKK RKVITGSVYR 240
241 WKSVIPFRFK GGDAKWKKLI REGLGLWEKE TCVRWSENGP GKDYVIFFRG SGCYSSVGRT 300
301 GGSQLISIGY GCEDKGIVAH EVGHSLGFWH EQSRPDRDDY IHLRKDWIIK GTDGNFEKRS 360
361 WEEIEDMGVP YDVGSVMHYG SNAFTKDWDQ ITIETKDSRY QGTIGQRQKL SFIDVKQVNR 420
421 LYCNSVCPVA LPCMHGGYPD PNNCAVCKCP DGLGGKLCGR AAKGTDHDKC GGELTATAEW 480
481 QEMVYKGKRT CNWKVKSPSG GRVRLVLTEL RYQCAPACKA YIEIKHNTDF QQTGFRVCCF 540
541 NKTYDVISDQ SEALILSNAN IVDYEVSYKL QWIQGTPPPP PFLSIERTFF QTTEKLFHHR 600
601 NPRRPGCPEK RIDHSAEWRT RAAPSKSSSC KRSQRSETRI GHWRVLLVLL RNMVSQRYLV 660
661 YRIMESSPFG TIFAIVRGIY IYICKFCTIP KIQFFFHNFQ NLLIFHNYSG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
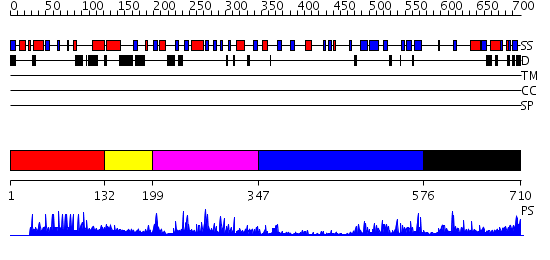
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [132..198] | 1.196995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [199..346] | 38.39794 | STRUCTURE OF ASTACIN AND IMPLICATIONS FOR ACTIVATION OF ASTACINS AND ZINC-LIGATION OF COLLAGENASES | |
| 4 | View Details | [347..575] | 23.221849 | Mannose-binding protein associated serine protease 2, MASP2 | |
| 5 | View Details | [576..710] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)