













| General Information: |
|
| Name(s) found: |
CE37093
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 765 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
signal transduction
[IEA protein amino acid phosphorylation [IEA |
| Molecular Function: |
ATP binding
[IEA protein tyrosine kinase activity [IEA non-membrane spanning protein tyrosine kinase activity [IEA protein kinase activity [IEA protein serine/threonine kinase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGNYKSRPLT SCSDELKKKI SEGYSVVRSR LSDDVRSRSN LGWVDVQIAA FEKSLEDFKQ 60
61 HMCPENAELK STQLLSLFHI ICAGHSDSQP EKLQFLIDNL PKESSITLIS SQSANGFTPL 120
121 HIAIYRGDVA ILKALIATKL VDLDQSGRHL LPALHLAAMI GDSEMLTILL NSGANIHVTD 180
181 FVHFTALHCA TYFGQENAVR TLISASANLN LGGAVNDRPI HLAAAKGLTS ITKLLLEAKA 240
241 DPLLADDEGN QALHYAAKSG SLVILNMLIK QVRGTNDRIC ARNLYGDTAL HLSCYSGRLD 300
301 IVKSILDSSP TNIVNMENVF SETPLHAACT GGKSIELVSF LMKYPGVDPN YQGQDGHTAL 360
361 HSACYHGHLR IVQYLLENGA DQSLASRAFE GGALRQQAGP GTNRPSKVAS AIMALNRSDT 420
421 PSSNASYNST VSLDDQQTPV IWAYERGHDA IVALLKHYAA RTVEGDVCSE YSSGESSYTP 480
481 LPSPMGRLTS LTRDKADLLQ LRSALPAPFH LCLAEIEFQE SIGSGSFGKV YKGTYRGKLV 540
541 AVKRYRAMAF GCKSETDMLC REVSILSRLA HPNVVAFVGT SLDDPSQFAI ITEFVENGSL 600
601 FRLLHEEKRV MDPAFRLRIS LDVARGMRYL HESAAKPVIH RDLNSHNILI HADGRSVVAD 660
661 FGESRFVCQR EDENLTKQPG NLRWMAPEVF SQSGKYDRKV DVFSFALVIW EIHTAELPFS 720
721 HRYKCSFYSI PVDISMGAAK RGTSPRVKVP ANSLGVTWDR CYGNR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
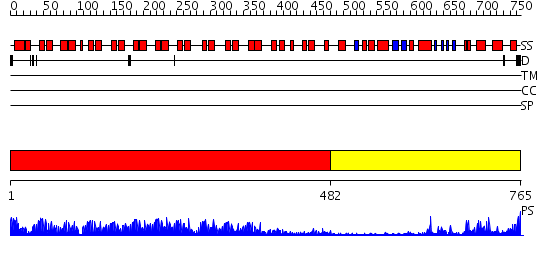
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..481] | 88.522879 | Ankyrin-R | |
| 2 | View Details | [482..765] | 27.09691 | Hemapoetic cell kinase Hck; Hemopoetic cell kinase Hck; Haemopoetic cell kinase Hck |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)