













| General Information: |
|
| Name(s) found: |
TRM2 /
YKR056W
[SGD]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 639 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
DNA repair
[IGI ribosome biogenesis [RCA tRNA modification [IDA |
| Molecular Function: |
double-stranded DNA specific 5'-3' exodeoxyribonuclease activity
[IDA tRNA (m5U54) methyltransferase activity [IDA single-stranded DNA specific endodeoxyribonuclease activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYEQFEFSFF FFENSDNKVK YKAHLISSIK RWSIITCMRC FWTVQKSIFK ARFFACRNFV 60
61 KKHNYKLIST MTGSTEMVPP TMKHTVDNKR LSSPLTDSGN RRTKKPKLRK YKAKKVETTS 120
121 PMGVLEFEVN DLLKSQNLSR EQVLNDVTSI LNDKSSTDGP IVLQYHREVK NVKVLEITSN 180
181 GNGLALIDNP VETEKKQVVI IPFGLPGDVV NIKVFKTHPY YVESDLLDVV EKSPMRRDDL 240
241 IRDKYFGKSS GSQLEFLTYD DQLELKRKTI MNAYKFFAPR LVAEKLLPPF DTTVASPLQF 300
301 GYRTKITPHF DMPKRKQKEL SVRPPLGFGQ KGRPQWRKDT LDIGGHGSIL DIDECVLATE 360
361 VLNKGLTNER RKFEQEFKNY KKGATILLRE NTTILDPSKP TLEQLTEEAS RDENGDISYV 420
421 EVEDKKNNVR LAKTCVTNPR QIVTEYVDGY TFNFSAGEFF QNNNSILPIV TKYVRDNLQA 480
481 PAKGDDNKTK FLVDAYCGSG LFSICSSKGV DKVIGVEISA DSVSFAEKNA KANGVENCRF 540
541 IVGKAEKLFE SIDTPSENTS VILDPPRKGC DELFLKQLAA YNPAKIIYIS CNVHSQARDV 600
601 EYFLKETENG SAHQIESIRG FDFFPQTHHV ESVCIMKRI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
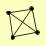
| View Details | Krogan NJ, et al. (2006) |
|
MNS1 RDH54 SIS2 TRM2 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
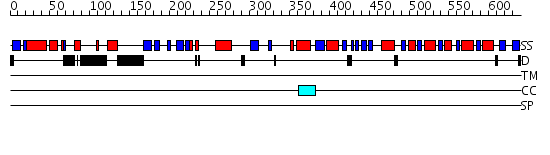
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..165] | 1.233938 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [166..639] | 77.221849 | Crystal Structure of RumA, the iron-sulfur cluster containing E. coli 23S Ribosomal RNA 5-Methyluridine Methyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)