













| General Information: |
|
| Name(s) found: |
FAT1 /
YBR041W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 669 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA peroxisome [IDA microsome [IDA plasma membrane [IMP |
| Biological Process: |
lipid transport
[IMP very long-chain fatty acid metabolic process [IMP |
| Molecular Function: |
long-chain fatty acid-CoA ligase activity
[IMP long-chain fatty acid transporter activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSPIQVVVFA LSRIFLLLFR LIKLIITPIQ KSLGYLFGNY FDELDRKYRY KEDWYIIPYF 60
61 LKSVFCYIID VRRHRFQNWY LFIKQVQQNG DHLAISYTRP MAEKGEFQLE TFTYIETYNI 120
121 VLRLSHILHF DYNVQAGDYV AIDCTNKPLF VFLWLSLWNI GAIPAFLNYN TKGTPLVHSL 180
181 KISNITQVFI DPDASNPIRE SEEEIKNALP DVKLNYLEEQ DLMHELLNSQ SPEFLQQDNV 240
241 RTPLGLTDFK PSMLIYTSGT TGLPKSAIMS WRKSSVGCQV FGHVLHMTNE STVFTAMPLF 300
301 HSTAALLGAC AILSHGGCLA LSHKFSASTF WKQVYLTGAT HIQYVGEVCR YLLHTPISKY 360
361 EKMHKVKVAY GNGLRPDIWQ DFRKRFNIEV IGEFYAATEA PFATTTFQKG DFGIGACRNY 420
421 GTIIQWFLSF QQTLVRMDPN DDSVIYRNSK GFCEVAPVGE PGEMLMRIFF PKKPETSFQG 480
481 YLGNAKETKS KVVRDVFRRG DAWYRCGDLL KADEYGLWYF LDRMGDTFRW KSENVSTTEV 540
541 EDQLTASNKE QYAQVLVVGI KVPKYEGRAG FAVIKLTDNS LDITAKTKLL NDSLSRLNLP 600
601 SYAMPLFVKF VDEIKMTDNH KILKKVYREQ KLPKGLDGND TIFWLKNYKR YEVLTAADWE 660
661 AIDAQTIKL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
CDC50 CSR1 DNF1 FAA1 FAA2 FAA4 FAT1 PDR16 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
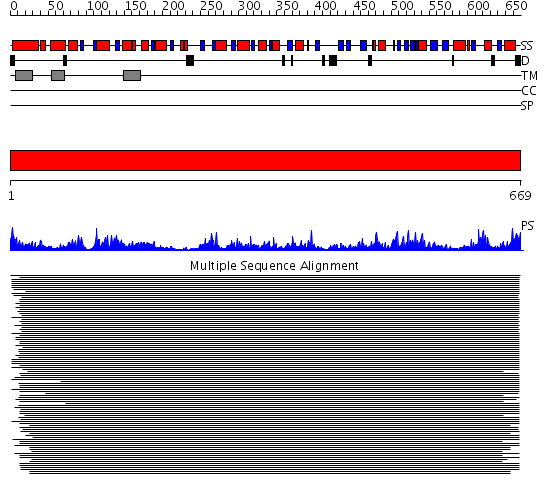
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..669] | 118.0 | Acetyl-CoA synthetase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.25 |
Source: Reynolds et al. (2008)