













| General Information: |
|
| Name(s) found: |
CDPK6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 544 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[TAS]
cytosol [IDA] plasma membrane [IDA] |
| Biological Process: |
abscisic acid mediated signaling pathway
[IMP]
regulation of stomatal movement [IMP] regulation of anion channel activity [IMP] protein amino acid phosphorylation [ISS] |
| Molecular Function: |
ATP binding
[IEA]
calcium ion binding [IEA] calmodulin-dependent protein kinase activity [ISS] protein kinase activity [IEA] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGNSCRGSFK DKIYEGNHSR PEENSKSTTT TVSSVHSPTT DQDFSKQNTN PALVIPVKEP 60
61 IMRRNVDNQS YYVLGHKTPN IRDLYTLSRK LGQGQFGTTY LCTDIATGVD YACKSISKRK 120
121 LISKEDVEDV RREIQIMHHL AGHKNIVTIK GAYEDPLYVH IVMELCAGGE LFDRIIHRGH 180
181 YSERKAAELT KIIVGVVEAC HSLGVMHRDL KPENFLLVNK DDDFSLKAID FGLSVFFKPG 240
241 QIFKDVVGSP YYVAPEVLLK HYGPEADVWT AGVILYILLS GVPPFWAETQ QGIFDAVLKG 300
301 YIDFDTDPWP VISDSAKDLI RKMLCSSPSE RLTAHEVLRH PWICENGVAP DRALDPAVLS 360
361 RLKQFSAMNK LKKMALKVIA ESLSEEEIAG LRAMFEAMDT DNSGAITFDE LKAGLRRYGS 420
421 TLKDTEIRDL MEAADVDNSG TIDYSEFIAA TIHLNKLERE EHLVSAFQYF DKDGSGYITI 480
481 DELQQSCIEH GMTDVFLEDI IKEVDQDNDG RIDYEEFVAM MQKGNAGVGR RTMKNSLNIS 540
541 MRDV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
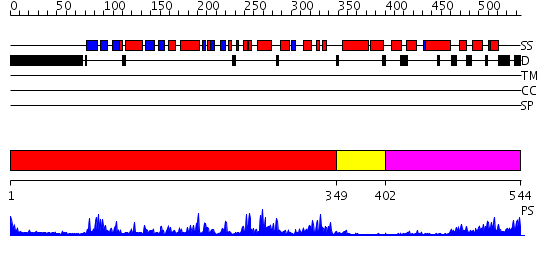
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..348] | 50.221849 | Abl tyrosine kinase, SH3 domain; Abl tyrosine kinase; Abelsone tyrosine kinase (abl) | |
| 2 | View Details | [349..401] | 70.69897 | No description for 2jc6A was found. | |
| 3 | View Details | [402..544] | 23.69897 | Ca2+-regulatory region (CLD) from soybean calcium-dependent protein kinase-alpha (CDPK) in the presence of Ca2+ and the junction domain (JD) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)