













| General Information: |
|
| Name(s) found: |
PDC1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 607 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IDA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
thiamin pyrophosphate binding
[IEA]
magnesium ion binding [IEA] pyruvate decarboxylase activity [ISS] catalytic activity [IEA] transferase activity [IEA] carboxy-lyase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTKIGSIDD CKPTNGDVCS PTNGTVATIH NSVPSSAITI NYCDATLGRH LARRLVQAGV 60
61 TDVFSVPGDF NLTLLDHLMA EPDLNLIGCC NELNAGYAAD GYARSRGVGA CVVTFTVGGL 120
121 SVLNAIAGAY SENLPLICIV GGPNSNDYGT NRILHHTIGL PDFSQELRCF QTVTCYQAVV 180
181 NNLDDAHEQI DKAISTALKE SKPVYISVSC NLAAIPHHTF SRDPVPFSLA PRLSNKMGLE 240
241 AAVEATLEFL NKAVKPVMVG GPKLRVAKAC DAFVELADAS GYALAMMPSA KGFVPEHHPH 300
301 FIGTYWGAVS TPFCSEIVES ADAYIFAGPI FNDYSSVGYS LLLKKEKAIV VQPDRITVAN 360
361 GPTFGCILMS DFFRELSKRV KRNETAYENY HRIFVPEGKP LKCESREPLR VNTMFQHIQK 420
421 MLSSETAVIA ETGDSWFNCQ KLKLPKGCGY EFQMQYGSIG WSVGATLGYA QASPEKRVLA 480
481 FIGDGSFQVT VQDISTMLRN GQKTIIFLIN NGGYTIEVEI HDGPYNVIKN WNYTGLVDAI 540
541 HNGEGNCWTA KVRYEEELVE AITTATTEKK DCLCFIEVIL HKDDTSKELL EWGSRVSAAN 600
601 SRPPNPQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
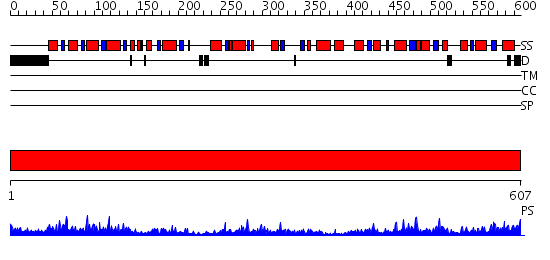
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..607] | 85.0 | Pyruvate decarboxylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.61 |
Source: Reynolds et al. (2008)