













| General Information: |
|
| Name(s) found: |
TAD2A_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 548 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to cold
[IMP]
|
| Molecular Function: |
DNA binding
[IEA][ISS]
zinc ion binding [IEA] transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGRSKLASRP AEEDLNPGKS KRKKISLGPE NAAASISTGI EAGNERKPGL YCCNYCDKDL 60
61 SGLVRFKCAV CMDFDLCVEC FSVGVELNRH KNSHPYRVMD NLSFSLVTSD WNADEEILLL 120
121 EAIATYGFGN WKEVADHVGS KTTTECIKHF NSAYMQSPCF PLPDLSHTIG KSKDELLAMS 180
181 KDSAVKTEIP AFVRLSPKEE LPVSAEIKHE ASGKVNEIDP PLSALAGVKK KGNVPQAKDI 240
241 IKLEAAKQQS DRSVGEKKLR LPGEKVPLVT ELYGYNLKRE EFEIEHDNDA EQLLADMEFK 300
301 DSDTDAEREQ KLQVLRIYSK RLDERKRRKE FVLERNLLYP DQYEMSLSAE ERKIYKSCKV 360
361 FARFQSKEEH KELIKKVIEE HQILRRIEDL QEARTAGCRT TSDANRFIEE KRKKEAEESM 420
421 LLRLNHGAPG SIAGKTLKSP RGLPRNLHPF GSDSLPKVTP PRIYSGLDTW DVDGLLGADL 480
481 LSETEKKMCN ETRILPVHYL KMLDILTREI KKGQIKKKSD AYSFFKVEPS KVDRVYDMLV 540
541 HKGIGDST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
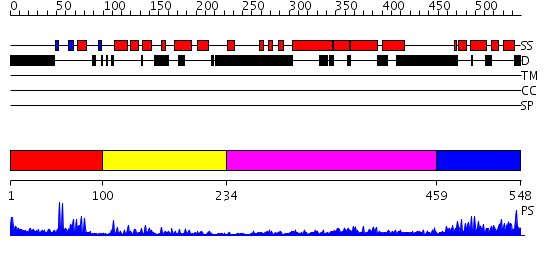
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | 6.522879 | No description for 2dipA was found. | |
| 2 | View Details | [100..233] | 17.69897 | c-Myb, DNA-binding domain | |
| 3 | View Details | [234..458] | 1.567994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [459..548] | 26.69897 | Solution structure of SWIRM domain of mouse transcriptional adaptor 2-like |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)