













| General Information: |
|
| Name(s) found: |
CDPKF_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 554 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
protein amino acid phosphorylation
[IEA][ISS]
|
| Molecular Function: |
ATP binding
[IEA]
calcium ion binding [IEA] protein kinase activity [IEA] calmodulin-dependent protein kinase activity [ISS] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGCFSSKHRN TESDIINGSV QSSIPTNQPE NHVSRDVLKP QKPPSPQIPT TTQSNHHHQQ 60
61 ESKPVNQQIE KKHVLTQPLK PIVFRETETI LGKPFEEIRK LYTLGKELGR GQFGITYTCK 120
121 ENSTGNTYAC KSILKRKLTR KQDIDDVKRE IQIMQYLSGQ ENIVEIKGAY EDRQSIHLVM 180
181 ELCGGSELFD RIIAQGHYSE KAAAGVIRSV LNVVQICHFM GVIHRDLKPE NFLLASTDEN 240
241 AMLKATDFGL SVFIEEGKVY RDIVGSAYYV APEVLRRSYG KEIDIWSAGI ILYILLCGVP 300
301 PFWSETEKGI FNEIIKGEID FDSQPWPSIS ESAKDLVRKL LTKDPKQRIS AAQALEHPWI 360
361 RGGEAPDKPI DSAVLSRMKQ FRAMNKLKKL ALKVIAESLS EEEIKGLKTM FANMDTDKSG 420
421 TITYEELKNG LAKLGSKLTE AEVKQLMEAA DVDGNGTIDY IEFISATMHR YRFDRDEHVF 480
481 KAFQYFDKDN SGFITMDELE SAMKEYGMGD EASIKEVIAE VDTDNDGRIN YEEFCAMMRS 540
541 GITLPQQGKI LPVQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
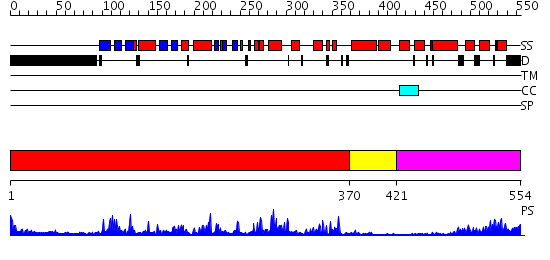
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..369] | 56.522879 | CHICKEN SRC TYROSINE KINASE | |
| 2 | View Details | [370..420] | 70.522879 | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | |
| 3 | View Details | [421..554] | 16.0 | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 1 STRUCTURE |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.63 |
Source: Reynolds et al. (2008)