













| General Information: |
|
| Name(s) found: |
SIR1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 473 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromatin silencing complex
[ISS]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[IEA][ISS]
protein amino acid deacetylation [IEA] chromatin silencing [IEA][ISS] |
| Molecular Function: |
DNA binding
[IEA][ISS]
zinc ion binding [IEA] NAD or NADH binding [IEA] NAD-dependent histone deacetylase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLGYAEKLS FIEDVGQVGM AEFFDPSHLL QCKIEELAKL IQKSKHLVVF TGAGISTSCG 60
61 IPDFRGPKGI WTLQREGKDL PKASLPFHRA MPSMTHMALV ELERAGILKF VISQNVDGLH 120
121 LRSGIPREKL SELHGDSFME MCPSCGAEYL RDFEVETIGL KETSRKCSVE KCGAKLKDTV 180
181 LDWEDALPPK EIDPAEKHCK KADLVLCLGT SLQITPACNL PLKCLKGGGK IVIVNLQKTP 240
241 KDKKANVVIH GLVDKVVAGV MESLNMKIPP YVRIDLFQII LTQSISGDQR FINWTLRVAS 300
301 VHGLTSQLPF IKSIEVSFSD NHNYKDAVLD KQPFLMKRRT ARNETFDIFF KVNYSDGCDC 360
361 VSTQLSLPFE FKISTEEHVE IIDKEAVLQS LREKAVEESS CGQSGVVERR VVSEPRSEAV 420
421 VYATVTSLRT YHSQQSLLAN GDLKWKLEGS GTSRKRSRTG KRKSKALAEE TKA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
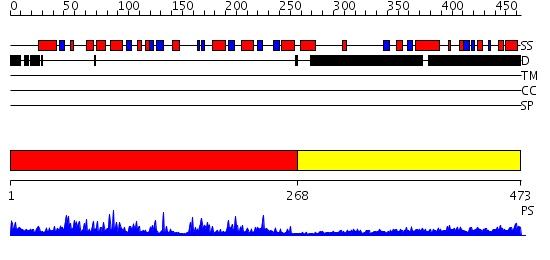
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..267] | 67.69897 | AF1676, Sir2 homolog (Sir2-AF1?) | |
| 2 | View Details | [268..473] | 20.157997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)