













| General Information: |
|
| Name(s) found: |
PDC2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 607 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to hypoxia
[IEP]
|
| Molecular Function: |
thiamin pyrophosphate binding
[IEA]
magnesium ion binding [IEA] pyruvate decarboxylase activity [ISS] catalytic activity [IEA] carboxy-lyase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTKIGSIDA CNPTNHDIGG PPNGGVSTVQ NTSPLHSTTV SPCDATLGRY LARRLVEIGV 60
61 TDVFSVPGDF NLTLLDHLIA EPNLKLIGCC NELNAGYAAD GYARSRGVGA CVVTFTVGGL 120
121 SVLNAIAGAY SENLPLICIV GGPNSNDYGT NRILHHTIGL PDFTQELRCF QAVTCFQAVI 180
181 NNLEEAHELI DTAISTALKE SKPVYISISC NLPAIPLPTF SRHPVPFMLP MKVSNQIGLD 240
241 AAVEAAAEFL NKAVKPVLVG GPKMRVAKAA DAFVELADAS GYGLAVMPSA KGQVPEHHKH 300
301 FIGTYWGAVS TAFCAEIVES ADAYLFAGPI FNDYSSVGYS LLLKKEKAII VQPDRVTIGN 360
361 GPAFGCVLMK DFLSELAKRI KHNNTSYENY HRIYVPEGKP LRDNPNESLR VNVLFQHIQN 420
421 MLSSESAVLA ETGDSWFNCQ KLKLPEGCGY EFQMQYGSIG WSVGATLGYA QAMPNRRVIA 480
481 CIGDGSFQVT AQDVSTMIRC GQKTIIFLIN NGGYTIEVEI HDGPYNVIKN WNYTAFVEAI 540
541 HNGEGKCWTA KVRCEEELVK AINTATNEEK ESFCFIEVIV HKDDTSKELL EWGSRVSAAN 600
601 SRPPNPQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
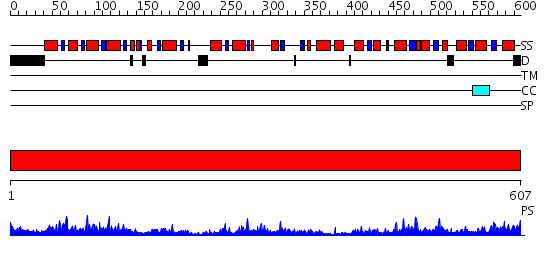
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..607] | 88.154902 | Pyruvate decarboxylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.61 |
Source: Reynolds et al. (2008)