













| General Information: |
|
| Name(s) found: |
HSD1B_ARATH
[Swiss-Prot]
HSD1A_ARATH [Swiss-Prot] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 349 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endomembrane system
[IEA]
|
| Biological Process: |
response to abscisic acid stimulus
[IMP]
metabolic process [IEA] response to brassinosteroid stimulus [IMP] |
| Molecular Function: |
oxidoreductase activity
[IEA][ISS]
catalytic activity [IEA] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELINDFLNL TAPFFTFFGL CFFLPPFYFF KFLQSIFSTI FSENLYGKVV LITGASSGIG 60
61 EQLAYEYACR GACLALTARR KNRLEEVAEI ARELGSPNVV TVHADVSKPD DCRRIVDDTI 120
121 THFGRLDHLV NNAGMTQISM FENIEDITRT KAVLDTNFWG SVYTTRAALP YLRQSNGKIV 180
181 AMSSSAAWLT APRMSFYNAS KAALLSFFET MRIELGGDVH ITIVTPGYIE SELTQGKYFS 240
241 GEGELIVNQD MRDVQVGPFP VASASGCAKS IVNGVCRKQR YVTEPSWFKV TYLWKVLCPE 300
301 LIEWGCRLLY MTGTGMSEDT ALNKRIMDIP GVRSTLYPES IRTPEIKSD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
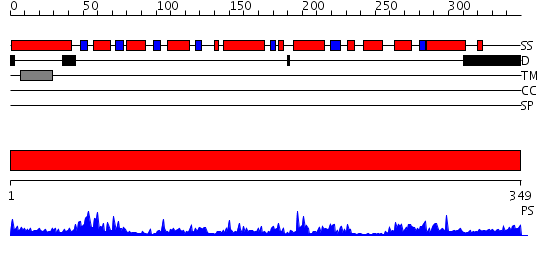
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..349] | 57.69897 | Crystal structure of peroxisomal trans 2-enoyl CoA reductase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|