













| General Information: |
|
| Name(s) found: |
MCM3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 776 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA replication initiation
[ISS]
DNA unwinding involved in replication [TAS] |
| Molecular Function: |
nucleotide binding
[IEA]
DNA binding [IEA][ISS] ATP binding [IEA][ISS] DNA-dependent ATPase activity [ISS] nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDVPEETRLR HKRDFIQFLD SMYMEEIKAL VHQKRHRLII NISDIHHHFR EVASRILKNP 60
61 NEYMQSFCDA ATEATRAIDP KYLKEGELVL VGFEGYFVSR VVTPRELLSD FIGSMVCVEG 120
121 IVTKCSLVRP KVVKSVHFCP STGEFTNRDY RDITSHAGLP TGSVYPTRDD KGNLLVTEYG 180
181 LCKYKDHQTL SIQEVPENAA PGQLPRSVDV IAEDDLVDSC KPGDRVSVFG IYKALPGKSK 240
241 GSVNGVFRTI LIANNIALLN KEANAPIYTK QDLDNIKNIA RRDDAFDLLA RSLAPSIYGH 300
301 AWIKKAVVLL MLGGVEKNLK NGTHLRGDIN MMMVGDPSVA KSQLLRAIMN IAPLAISTTG 360
361 RGSSGVGLTA AVTSDQETGE RRLEAGAMVL ADKGIVCIDE FDKMNDQDRV AIHEVMEQQT 420
421 VTIAKAGIHA SLNARCSVVA AANPIYGTYD RSLTPTKNIG LPDSLLSRFD LLFIVLDQMD 480
481 AGIDSMISEH VLRMHRYKND RGEAGPDGSL PYAREDNAES EMFVKYNQTL HGKKKRGQTH 540
541 DKTLTIKFLK KYIHYAKHRI TPKLTDEASE RIAEAYADLR NAGSDTKTGG TLPITARTLE 600
601 TIIRLATAHA KMKLSSEVTK ADAEAALKLM NFAIYHQELT EMDDREQEER QREQAEQERT 660
661 PSGRRGNQRR NNEDGAENDT ANVDSETADP MEVDEPSVEQ FSGTVSAARI ETFERVFGQH 720
721 MRTHRLDDIS IADIETVVNN NGVGASRYSA DEIMALLEKL QDDNKVMISD GKVHII |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
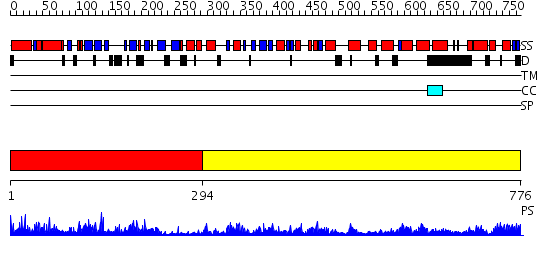
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..293] | 45.045757 | DNA replication initiator (cdc21/cdc54) N-terminal domain | |
| 2 | View Details | [294..776] | 75.69897 | No description for 2dhrA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)