













| General Information: |
|
| Name(s) found: |
gi|5732432
[NCBI NR]
gi|15241729 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 625 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
mRNA capping
[IEA][ISS]
protein amino acid dephosphorylation [IEA] mRNA processing [IEA][ISS] dephosphorylation [IEA] |
| Molecular Function: |
protein tyrosine phosphatase activity
[IEA]
phosphatase activity [IEA] mRNA guanylyltransferase activity [IEA][ISS] polynucleotide 5'-phosphatase activity [IEA] protein tyrosine/serine/threonine phosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MISTMYSEET SERSSFPSKR HLEDRTENRE EHSKRLRPTS FRDQKVTIPQ GWLDCPRFGQ 60
61 HIGLIIPSKV PLSESYNDCV PSGKRYNFKQ WLTKIGLVID LTNTTRYYHP NTELRQNRIE 120
121 YVKIRCSGRD SVPDNVSVNT FVHEVTQFEN HNLSEKYLLV HCTHGHNRTG FMIVHYLMRS 180
181 RPMMSVTQAL KIFSDARPPG IYKPDYIDAL YRFYHEVKPG SVICPPTPEW KRSEEAKFDD 240
241 DDNALSYREV QGNNQEENVQ LSNDDVLGDE IPYDQEVSYQ NSINHMLNIS MQFPGSHPLS 300
301 LGREALQLLR QRYYYATWKA DGTRYMMLLT RDGCYLVNRE YRFRRVQMRF PCEYEPSDYK 360
361 VHHYTLLDGE MVVDTIKEGE TQRHVRRYLV YDLVAINGQF VAERPFSERW NILERELIKP 420
421 RNDEKKVRDH WYRYDKEPFG VRIKAFCLLS AVEKKVFNEL IPLLSHESDG LIFQGWDNPY 480
481 VFGPNKDLLK WKFVETLDFL FDMDKYGRQM LFLQERGRMK LMEGFAVEFR GDGWDNDLAS 540
541 YCGKIVECSW DKEKKVWVSL RIRVDKSKPN GIGTGRSVIK CIEDDLTKEV LLKEIKEIIL 600
601 LPMYVERIEN DTKEAARRKH GNRKG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
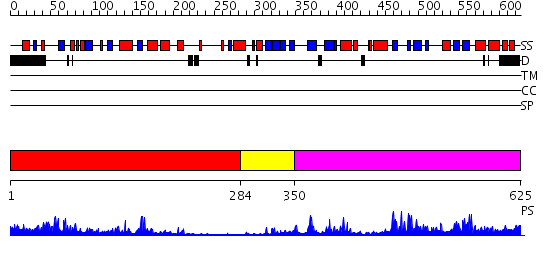
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..283] | 23.522879 | CRYSTAL STRUCTURE OF THE HUMAN RNA guanylyltransferase and 5'- phosphatase | |
| 2 | View Details | [284..349] | 9.39794 | Crystal structure of the m3G-cap-binding domain of snurportin1 in complex with a m3GpppG-cap dinucleotide | |
| 3 | View Details | [350..625] | 39.0 | mRNA capping enzyme alpha subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)