













| General Information: |
|
| Name(s) found: |
CSN1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 441 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS]
signalosome [IPI][TAS] |
| Biological Process: |
photomorphogenesis
[TAS]
response to salt stress [IEP] protein complex assembly [IDA] embryonic development ending in seed dormancy [IMP] protein deneddylation [TAS] hyperosmotic response [IEP] response to absence of light [IEP] cullin deneddylation [IMP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERDEEASGP MMEMCTNGGE ETSNRRPIIS GEPLDIEAYA ALYKGRTKIM RLLFIANHCG 60
61 GNHALQFDAL RMAYDEIKKG ENTQLFREVV NKIGNRLGEK YGMDLAWCEA VDRRAEQKKV 120
121 KLENELSSYR TNLIKESIRM GYNDFGDFYY ACGMLGDAFK NYIRTRDYCT TTKHIIHMCM 180
181 NAILVSIEMG QFTHVTSYVN KAEQNPETLE PMVNAKLRCA SGLAHLELKK YKLAARKFLD 240
241 VNPELGNSYN EVIAPQDIAT YGGLCALASF DRSELKQKVI DNINFRNFLE LVPDVRELIN 300
301 DFYSSRYASC LEYLASLKSN LLLDIHLHDH VDTLYDQIRK KALIQYTLPF VSVDLSRMAD 360
361 AFKTSVSGLE KELEALITDN QIQARIDSHN KILYARHADQ RNATFQKVLQ MGNEFDRDVR 420
421 AMLLRANLLK HEYHARSARK L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
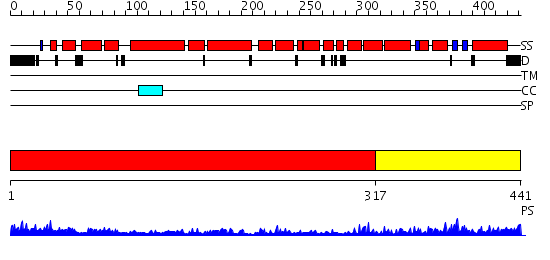
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..316] | 3.08 | No description for 2gw1A was found. | |
| 2 | View Details | [317..441] | 9.0 | Solution structure of the PCI domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)