













| General Information: |
|
| Name(s) found: |
cul-3 /
CE27593
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 777 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cullin-RING ubiquitin ligase complex
[IEA |
| Biological Process: |
pronuclear migration
[IMP embryonic development ending in birth or egg hatching [IMP cell cycle [IEA |
| Molecular Function: |
ubiquitin protein ligase binding
[IEA protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGRSGNGGQ QKMRIRPFMA TIDEQYVTQT WELLKRAIQE IQRKNNSGLS FEELYRNAYT 60
61 MVLHKHGERL YNGLKDVIQD HMASVRIRII ESMNSGSFLE TVAESWADHT VAMVMIRDIL 120
121 MYMDRIYVAQ NNHVLPVYNL GLDAYRTEIL RQNGIGDRIR DALLELIKLD RKSNQINWHG 180
181 IKNACDMLIS LGIDSRTVYE DEFERPLLKE TSDYYRDVCK NWLSGDNDAC FYLAQVEIAM 240
241 HDEASRASRY LDKMTEAKIL QVMDDVMVAE HIQTIVYMQN GGVKFMLEHK KIEDLTRIFR 300
301 IFKRIGDSVT VPGGGLKALL KAVSEYLNET GSNIVKNEDL LKNPVNFVNE LLQLKDYFSS 360
361 LLTTAFADDR DFKNRFQHDF ETFLNSNRQS PEFVALYMDD MLRSGLKCVS DAEMDNKLDN 420
421 VMILFRYLQE KDVFEKYFKQ YLAKRLLLDK SCSDDVEKAL LAKLKTECGC QFTQKLENMF 480
481 RDKELWLTLA TSFRDWREAQ PTKMSIDISL RVLTAGVWPT VQCNPVVLPQ ELSVAYEMFT 540
541 QYYTEKHTGR KLTINTLLGN ADVKATFYPP PKASMSNEEN GPGPSSSGES MKERKPEHKI 600
601 LQVNTHQMII LLQFNHHNRI SCQQLMDELK IPERELKRNL QSLALGKASQ RILVRKNKGK 660
661 DAIDMSDEFA VNDNFQSKLT RVKVQMVTGK VESEPEIRET RQKVEDDRKL EVEAAIVRIM 720
721 KARKKLNHNN LVAEVTQQLR HRFMPSPIII KQRIETLIER EYLARDEHDH RAYQYIA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
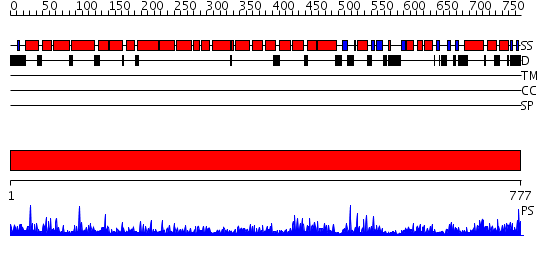
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..777] | 1000.0 | No description for 2hyeC was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)