













| General Information: |
|
| Name(s) found: |
tufm-1 /
CE27322
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 496 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA intracellular [IEA |
| Biological Process: |
selenocysteine incorporation
[IEA nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP translational elongation [IEA growth [IMP reproduction [IMP |
| Molecular Function: |
GTP binding
[IEA RNA binding [IEA translation elongation factor activity [IEA GTPase activity [IEA transferase activity, transferring phosphorus-containing groups [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQILNLLSRN VGVFKTVTLC EQKMQQVSLM QARHLAVPGG KAVFKRDKPH LNVGTIGHVD 60
61 HGKTTLTSAI TKILATSKGA KYRKYEDIDN APEEKARGIT INAFHLEYET AKRHYAHIDC 120
121 PGHADYIKNM ITGAAQMEGA ILVVAATDGP MPQTREHLLL ARQVGVPLDN IVVFMNKVDE 180
181 VPDAETRELV EMDIREQLNE FGYPGDTCPV IFGSALCALE GKQPEIGEEA VKQLLEVLDN 240
241 KFVIPERKVN EEPMFAAEHV YSIVGRGTVI TGKLERGILK RGDKIEIVGG TKDGTTVKSV 300
301 ISGLESFRKT VDQAEPGDQL GVLLRGLGPK DVRRGCVLLP QGHKHKVTDK VKAQLYVLKE 360
361 SEGGAKTPIA NYFSEHVFSL TWDSGASVRI IGKDFVMPGE SAEVELSLNS QMFIEPQQRF 420
421 TIRKGAKTIG TGVFTDVLPP LTNDEKDPKS KKKLMKAEME RLGFNPYGDM VEKRLKPDFS 480
481 NSPKDNPAAK AFEGQP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
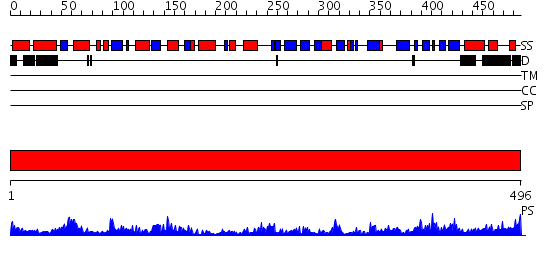
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..496] | 98.0 | Crystal Structure of Bos taurus mitochondrial Elongation Factor Tu/Ts Complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)