













| General Information: |
|
| Name(s) found: |
dpy-31 /
CE27200
[WormBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 592 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP proteolysis [IEA |
| Molecular Function: |
meprin A activity
[IEA zinc ion binding [IEA astacin activity [IEA metallopeptidase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHKIFIIFGL LSLCAAHSLR DLSNKDEEDP PSSAPGVRKR RMMSEEDQKT VDYYMDKLNK 60
61 LADEKHPEEI ERHKNPELVA WDRKRDSVLN PEEQGKFFQG DIVLYPEQAK ALYEQALTEG 120
121 KTRVKRKFIG SNLRRWDASR PIIYAFDGSH TQREQRIIEL ALEHWHNITC LNFQRNDQAN 180
181 SGNRIVFTDV DGCASNVGRH PLGEEQLVSL APECIRLGVI AHEVAHALGF WHEQSRPDRD 240
241 QYVTVRWENI DKDSKGQFLK EDPDDVDNAG VPYDYGSIMH YRSKAFSKFD DLYTISTYVT 300
301 DYQKTIGQRD QLSFNDIRLM NKIYCSAVCP SKLPCQRGGY TDPRRCDRCR CPDGFTGQYC 360
361 EQVMPGYGAT CGGKISLTRS TTRISSPGYP REFKEGQECS WLLVAPPGHI VEFQFIGEFE 420
421 MYCKIRHSLC MDYVEVRNST DFANTGMRYC CYGTPPTRIR SATTDMVVLF RSFYRGGKGF 480
481 EARARAVPEA GNWNSWSPWT ACSATCGACG SRMRTRTCPP GNACSGEPVE TQICNTQACT 540
541 GMCAQKREEE GQCGGFLSLL RGVRCRQEKT VMAPCENACC PGFTLQRGRC VR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
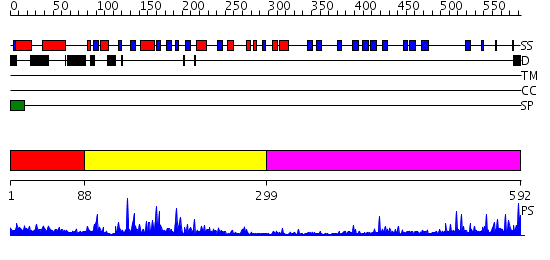
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [88..298] | 34.221849 | STRUCTURE OF ASTACIN AND IMPLICATIONS FOR ACTIVATION OF ASTACINS AND ZINC-LIGATION OF COLLAGENASES | |
| 3 | View Details | [299..592] | 18.522879 | Low density lipoprotein (LDL) receptor YWTD domain; Low density lipoprotein (LDL) receptor, different EGF domains; Ligand-binding domain of low-density lipoprotein receptor |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|