













| General Information: |
|
| Name(s) found: |
CE26950
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 755 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fatty acid beta-oxidation multienzyme complex
[IEA mitochondrion [IEA |
| Biological Process: |
fatty acid catabolic process
[IEA fatty acid beta-oxidation [IEA nematode larval development [IMP metabolic process [IEA valine metabolic process [IEA menaquinone biosynthetic process [IEA fatty acid metabolic process [IEA growth [IMP |
| Molecular Function: |
naphthoate synthase activity
[IEA catalytic activity [IEA oxidoreductase activity [IEA 3-hydroxyacyl-CoA dehydrogenase activity [IEA NAD or NADH binding [IEA 3-hydroxybutyryl-CoA epimerase activity [IEA 3-hydroxyisobutyrate dehydrogenase activity [IEA dodecenoyl-CoA delta-isomerase activity [IEA enoyl-CoA hydratase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSRLVHQSS STLRTNLSFR LFSQSAPAMQ TTHRVEKQGD VAVVKIDLPN TKENVLNKAL 60
61 FAEMKATLDK LQSDESIKSI VVMSGKPNSF VAGADIQMIK AEGTATATET LSREGQEQFF 120
121 RIEKSQKPVV AAIMGSCMGG GLELALACHY RIAVNDKKTL LSLPEVMLGL LPGAGGTQRL 180
181 PKLTTVQNVL DLTLTGKKIK ADKAKKIGIV DRVIQPLGDG LGPAAENTHK YLEEIAVKAA 240
241 QELANGKLKI NRDKGFMHKA TQAVMTNSLF LDNVVLKMAK DKLMKLTAGN YPAPLKILDV 300
301 VRTAYVDPKK GFEAEAKAFG ELSQTFQSKA LIGLFDGSTD AKKNKYGKGL PVNEIAVVGA 360
361 GLMGAGIANV TIDKGIRTVL LDANPAGVER GQNQIATHLN KQVKRRKINK LEKERIYNHL 420
421 VPTIDYSAMK NADVVIEAVF EDLQLKHKVI KQIESVVGPN TIIASNTSAL PIKDIAAASS 480
481 RPDKVIGMHY FSPVEKMQLL EIITHDGTSK ETLATAAQLG LKQGKLVVVV KDCPGFFVVR 540
541 CLGPMTAEIM RLLQEGVDPA ELDKLTTKFG FPVGAATLAD EVGLDVAEHV AQFLGKALGP 600
601 RLGGGSADLL SELVKAGHKG RKTGKGIFVY GDGKKGSKKV NDEATKLLQK YKLTPNASVS 660
661 SPEDRQIRLV SRFVNEALLC LEEGVLASPS DGDIASVFGV GFPPFWGGPF RFVDLYGADK 720
721 LVKSMEKFAG AYEAVQFEPC QLLKDHAKSG KKFYN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
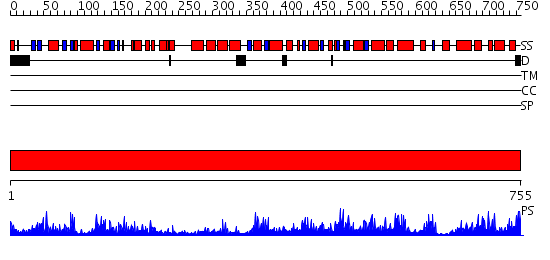
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..755] | 177.0 | fatty acid beta-oxidation multienzyme complex from Pseudomonas fragi, form I (native2) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)