













| General Information: |
|
| Name(s) found: |
CE17558
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 473 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
electron transport
[IEA removal of superoxide radicals [IEA glutathione metabolic process [IEA cell redox homeostasis [IEA detoxification of mercury ion [IEA |
| Molecular Function: |
disulfide oxidoreductase activity
[IEA NADP or NADPH binding [IEA oxidoreductase activity [IEA FAD binding [IEA mercury ion binding [IEA mercury (II) reductase activity [IEA thioredoxin-disulfide reductase activity [IEA glutathione-disulfide reductase activity [IEA dihydrolipoyl dehydrogenase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRFRCILST SRSIMSGVKE FDYLVIGGGS GGIASARRAR EFGVSVGLIE SGRLGGTCVN 60
61 VGCVPKKVMY NCSLHAEFIR DHADYGFDVT LNKFDWKVIK KSRDEYIKRL NGLYESGLKG 120
121 SSVEYIRGRA TFAEDGTVEV NGAKYRGKNT LIAVGGKPTI PNIKGAEHGI DSDGFFDLED 180
181 LPSRTVVVGA GYIAVEIAGV LANLGSDTHL LIRYDKVLRT FDKMLSDELT ADMDEETNPL 240
241 HLHKNTQVTE VIKGDDGLLT IKTTTGVIEK VQTLIWAIGR DPLTKELNLE RVGVKTDKSG 300
301 HIIVDEYQNT SAPGILSVGD DTGKFLLTPV AIAAGRRLSH RLFNGETDNK LTYENIATVV 360
361 FSHPLIGTVG LTEAEAVEKY GKDEVTLYKS RFNPMLFAVT KHKEKAAMKL VCVGKDEKVV 420
421 GVHVFGVGSD EMLQGFAVAV TMGATKKQFD QTVAIHPTSA EELVTMRGGV KPE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
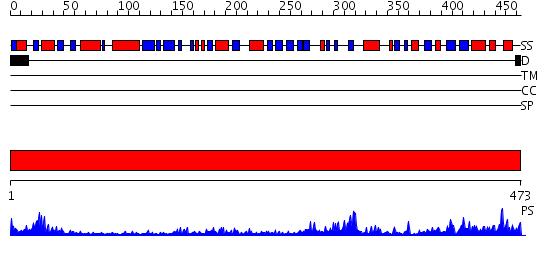
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..473] | 100.0 | HUMAN GLUTATHIONE REDUCTASE A34E/R37W MUTANT |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)