













| General Information: |
|
| Name(s) found: |
cul-6 /
CE11928
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 729 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cullin-RING ubiquitin ligase complex
[IEA |
| Biological Process: |
DNA repair
[IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP cell cycle [IEA |
| Molecular Function: |
ubiquitin protein ligase binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIEAVWGTL QDGLNLLYRR EHMSKKYYMM LYDAVYNICT TTTLANSNNN SPEFASEFLY 60
61 KQLENYIRTY VIAIRDRISA CSGDELLGKC TIEWDNFKFS TRICNCIFQY LNRNFVSKKV 120
121 EDKNGEIVEI YKLALDIWKA EFFDNFKVKT IDAILELILL ERCGSTINST HISSVVECLT 180
181 ELDIYKVSFE PQFLDATKLF YKQEVLNSKE TVIEYMITVE NRLFQEEYRS RRYLGPSTND 240
241 LLIDSCESIL ISDRLKFLHS EFERLLEARK DEHLTRMYSL CRRVTHGLED LRVYLEKRIL 300
301 KEGHETLQRL AKDSGLKTTP KEYITKLLEV HEIYFNLINK AFDRNALFMQ SLDKASKDFI 360
361 EANAVTMLAP EKHRSTRSAD YLARYCDQLL KKNSKVQDET ALDKALTVLK YISEKDVFQL 420
421 YYQNWFSERI INNSSASDDA EEKFITNLTA TEGLEYTRNL VKMVEDAKIS KDLTTEFKDI 480
481 KTEKSIDFNV ILQTTGAWPS LDQIKIILPR ELSTILKEFD TFYNASHNGR RLNWAYSQCR 540
541 GEVNSKAFEK KYVFIVTASQ LCTLYLFNEQ DSFTIEQISK AIEMTAKSTS AIVGSLNPVI 600
601 DPVLVVDKGN EKDGYPPDAV VSLNTKYANK KVRVDLTTAI KKATADRETD AVQNTVESDR 660
661 KYEIKACIVR IMKTRKSLTH TLLINEIISQ LKSRFTPNVQ MIKICIEILI EQLYIRRSEN 720
721 EHNVYEYLA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
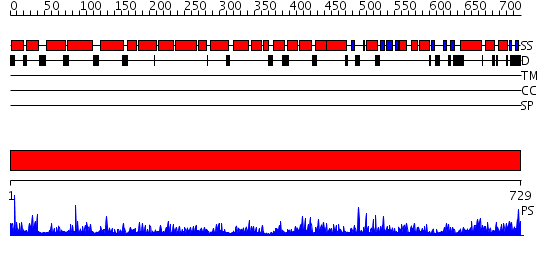
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..729] | 1000.0 | Crystal Structure of The Cand1-Cul1-Roc1 Complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)