













| General Information: |
|
| Name(s) found: |
alh-11 /
CE07233
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 687 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IEA cytoplasm [IEA |
| Biological Process: |
cellular aldehyde metabolic process
[IEA proline metabolic process [IEA negative regulation of transcription [IEA metabolic process [IEA 10-formyltetrahydrofolate catabolic process [IEA proline biosynthetic process [IEA valine metabolic process [IEA betaine biosynthetic process [IEA one-carbon metabolic process [IEA |
| Molecular Function: |
1-pyrroline-5-carboxylate dehydrogenase activity
[IEA DNA binding [IEA NADP or NADPH binding [IEA betaine-aldehyde dehydrogenase activity [IEA proline dehydrogenase activity [IEA oxidoreductase activity [IEA aldehyde dehydrogenase [NAD(P)+] activity [IEA methylmalonate-semialdehyde dehydrogenase (acylating) activity [IEA glutamate-5-semialdehyde dehydrogenase activity [IEA formyltetrahydrofolate dehydrogenase activity [IEA transcription repressor activity [IEA oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLKPGDVVFY SRGTESTDFC DAVEKVVPDN TYYHVAIVSE RGTIVEATGD GVLEYSLQKS 60
61 IEENRPGCLE ILEVANCCEK VLEAAATWSW TKLGLPYNDL FAADLINSDG KESYYCSQLI 120
121 TAAFEFAGVD MKWPAHTLNF NDKNVNQISF WNEYFEKRGK SQVPQGEEGS HPAQLRRSPR 180
181 LLLKMRIFPN LINLNSLTET KLLELSSHFV AGNHVEFPSD RKFEVIEPRS GKPMATWHYA 240
241 TRDQVDLTVK EAKKAQKQWA KSSWMERSEI LKKTGDLLKT HCNDIAYWEC ISNGKPIAEA 300
301 KADVLSCVDT FYFYSGIASD LLGQHVPLDA SRYAYTRRLP VGVVAAIGAW NYPIQTCSWK 360
361 TAPALACGNA VIYKPSPLSP VTALILAEIL KSAGLPDGVF NVIQGDAETA QDLIHHDGVS 420
421 KVSFTGSIPT GKKIMKACAD RNIKPVTLEL GGKSALIVFD DADIDSAVSC AMMANFYSQG 480
481 QVCSNASKVL VHKSVLKEFT KKLVDHTQKM KIGDPLKEDT QVGSHISAEH RNKVEGYIST 540
541 AIAEGATKLC GGDRVAVHGL ENGFYLSPCI LTGITPKMTV YREEIFGSVL LIIPFDTEDE 600
601 AIKIANDTDM GLAAGLVTKD LSRSYRVSEQ LNAGNVYVNT YNDVSPLVPF GGVGESGFGR 660
661 ENGTAVLEHY THLKSVFVNT GTCPNPF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
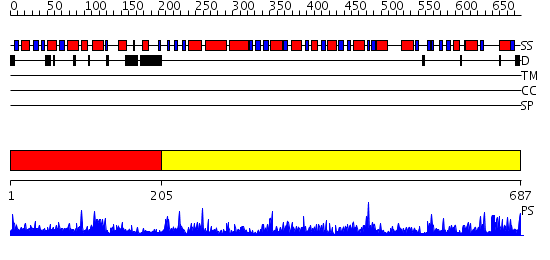
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..204] | 39.522879 | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with acetate | |
| 2 | View Details | [205..687] | 155.0 | Aldehyde reductase (dehydrogenase), ALDH |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)