













| General Information: |
|
| Name(s) found: |
SPCC11E10.06c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 361 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [ISS Elongator holoenzyme complex [ISS cytosol [IDA |
| Biological Process: |
tRNA modification
[ISS regulation of transcription from RNA polymerase II promoter [IEA] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFKRKAAPQ IAPTNLPTGV RLSSKDARWI TSSGSSSFDY YLSGGIPMKS LLVIEEDSMD 60
61 YASVLLKFFA AEGLKQDHVI WLGPSIGEMW FRQLPGDSDR PNKNENSAGE DNHSSPPSKN 120
121 PQQERMKIAW RYEQVSKTKA PTLDMIPPGY THSFDLSKNL IVKSDMKYAV SPFPLETGSN 180
181 PYAPVIESLT RFLSTLTPGT VCRLVLPSIL SPAFYSIRAT HPQHFIRFIH TLSSLIKCTT 240
241 SVHLICMCSV PSTLFSRDCE QIFWLENLAS AVFSLHPFPV KETVNGLVTQ PLGLFRIHKL 300
301 PLALPFTNHA NSNEAGDLSF TVSKRRFTIE PWVLPPLDDE QKDTKISNTN PQKQPVKSLD 360
361 F |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
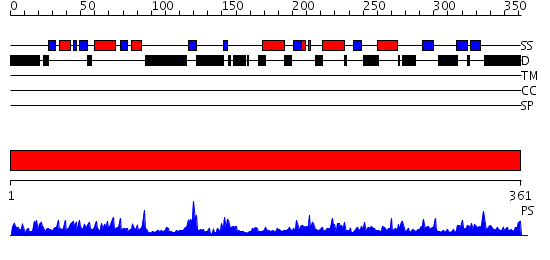
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..361] | 2.44 | Crystal Structure of E. Coli RecA in a Compressed Helical Filament Form 2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)