













| General Information: |
|
| Name(s) found: |
SPBC2A9.04c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 741 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
chromatin silencing
[ISS chromatin organization [ISS ubiquitin-dependent protein catabolic process [ISS protein ubiquitination [IC |
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS zinc ion binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSNTNENNS HASSNERQSS EGHDDYLNRN PNSEATEGEE GTHPTTGTQP VAFSIGTMFI 60
61 ITPQANFEGG ENDPFANPFQ PPVKRAVKEA WDSFEPLSND QLMDLTCPIC YDDMNENDEK 120
121 QATKMPCGHI FGKNCLQKWL ENHCTCPLCR KEVPHETVGS AHPPILFIIP HSHTLRGNQG 180
181 NTAVSQENAS NGVHSDFHPS EELNNANTDG RTGVDEPARQ LHRIAFNRIR FILAPNRSAT 240
241 NTPVENTHPE NPDSNTSTPT TRSEPLAGEG ASIDAENASS RQETTPSDSR PSTLTSLFNA 300
301 FFSSMPDRPS SNEPMTSNLT SNSGSMTNST STDLPTSNLP SQNAPARPVE PSPSIQPPNL 360
361 LNLPTASPES TSWLPGSQTN IPANTNRSER PFTQLMTFHG LPSLADLPAV LESMFRPSGN 420
421 NNLLNLNGIF HPDHNAQTEN GQTLPENTDD TNSNATSAVP NLQNLNQQNA VTMGTNTPNN 480
481 GSSPAVHPVI HIYLSRPPLQ PAVSEQETPS EAVSREGTRS TDATMEDSSR PPSNGSFQGP 540
541 GITHLSEMGQ RILQRFQEEM ENRMNQTRSE SSTPAQQSAA GSSINVDTAG RQPSDEINIP 600
601 GEYENSSEVA GSRNQTPTHS SIAVDTLSNV AVDQPAISTP SDVVGSDAGD TSKVSSGTST 660
661 PRMAAPIARR SNRHHPYSRP SSTRPQCQLE DQGICDPNDR FVHFECGHSV HERCQQSTSN 720
721 SENQMDEEIG ECPKCRNEEH K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
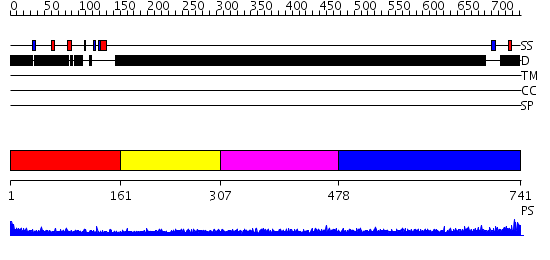
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..160] | 3.17 | Crystal Structure of The Cand1-Cul1-Roc1 Complex | |
| 2 | View Details | [161..306] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [307..477] | 2.194989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [478..741] | 3.200995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)