













| General Information: |
|
| Name(s) found: |
sap61 /
SPBC36.09
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 492 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spliceosomal complex [IDA cytosol [IDA U2 snRNP [ISS |
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[ISS |
| Molecular Function: |
zinc ion binding
[IEA]
RNA binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSESVLETER YAHEELERLQ QAIVDRQVAN PKAPRERLRL EHQSAQFLNQ FRETSKKLLV 60
61 SHESSDRLKD QEVARINADD DLTEFYKSLG EIQEFHKKYP DHKVEDLSQL YSIKPSQPGI 120
121 DEIDTLFRGE EMYGRFMDLN ECYEEYINLS NVQHISYLEY LKNLEDFDQI PKPEKNQTYI 180
181 NYITHLYEYL VSFYRRTHPL SNLDKIIAVF DTEFDAAWEA GLPGWYSHNA EAEKDGKDST 240
241 EAFYCEVCQK FFGKITVFEA HKKSKAHNKA VKRMQSSSPS TTSNTNEKQK GPKAIARIEF 300
301 LIKKLTSLLD DVRKDTRENV VRRQTLTAAE RLAEVEAAER EAFEQSTPSV SVEGNQDEES 360
361 DQDDEEKIYN PLKLPLGWDG KPIPFWLWKL HGLGKEFPCE ICGNYVYMGR KAFDKHFTEQ 420
421 RHIYGLKCLG ISPSPLFNQI TSIDEALQLW QKYKVDSKKR ETTMASLNEM EDDEGNVMSE 480
481 KVYNDLKAQG LL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
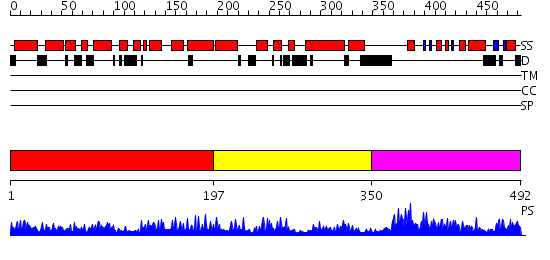
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | 1.037955 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [197..349] | 2.15 | Solution Structure of a Human C2H2-type Zinc Finger Protein | |
| 3 | View Details | [350..492] | 2.042961 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)