













| General Information: |
|
| Name(s) found: |
hst4 /
SPAC1783.04c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 415 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IC chromatin remodeling complex [ISS nucleolus [IDA |
| Biological Process: |
DNA damage checkpoint
[IMP chromatin remodeling [ISS chromatin silencing at centromere [IMP mitotic sister chromatid segregation [IMP histone deacetylation [IMP response to DNA damage stimulus [IMP chromatin silencing at telomere [IMP regulation of nucleosome density [IEP negative regulation of gene-specific transcription from RNA polymerase II promoter [IEP histone H3 deacetylation [IMP |
| Molecular Function: |
zinc ion binding
[IEA]
transcription corepressor activity [IEP transcription regulator activity [IC NAD-dependent histone deacetylase activity [TAS specific transcriptional repressor activity [IEP NAD binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKVEEHVPLI QESRKRKCQS SENASKRQQL LSKLPLRLHT GNENVDLSPL VSAIRKAKRI 60
61 VVVTGAGISC DAGIPDFRSS EGLFSSLRAE YKLNCSGKEL FDGSVYRDLK SVNIFHAMIR 120
121 KLHMLSNNAR PTDFHLFLSQ LAQESKLLRL YTQNIDFLET RLEGLQTCIP LPQSAPWPTT 180
181 IPLHGTLEVV SCTRCSFLKK FNPDIFDRNG VTVCPDCKTE NEVRRIAGKR SVIEGCLRPR 240
241 IVLYNEIHPD SESIGSVCSQ DLKSRPDCLI VAGTSCKIPG VKRIIKEMSN CVHKQKGNVI 300
301 WLNYDEPTKD FLNLCDLVVQ GDLQIAIRRL KPLLDAPSWK LKSHSAKRTS KQKSSEQTKI 360
361 TSSTKITKAI GLNTKSNDSS KKDNTSFQLH QVLNSIEIPK VEIKQEVEYA TPSPL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
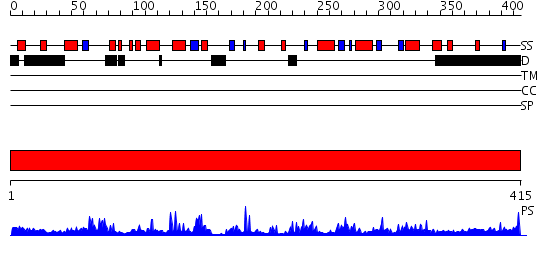
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..415] | 64.221849 | Structure and autoregulation of the yeast Hst2 homolog of Sir2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)