













| General Information: |
|
| Name(s) found: |
rhp26 /
SPCP25A2.02c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 973 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
transcription-coupled nucleotide-excision repair
[IMP |
| Molecular Function: |
ATP binding
[ISS DNA binding [IEA] DNA-dependent ATPase activity [ISS ATP-dependent DNA helicase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVNEDLSHL GVFSVDQENL ERDVTNTASE YIAHESREIE KKRLQKVRKE ISSVKEKIRR 60
61 LDERIDSRLT KISVKENFRK QLSKFRDTLQ SLQSDENDIK RRLNNEDSAN APGIGAFSTE 120
121 ELERQELIRT GKVTPFRNLS GLQKEVDFDD ESSIREAVIK SEGTYYETAP HLSSEPSNID 180
181 HGIIPRDEKD EYVTVDAVTE KVVTAAIDDG DDLVYRQRLN AWCANRKELR DQASASENNK 240
241 DRGEFEGKDE WLLPHPSKKG QTFEGGFTIP GDIRPHLFRY QVTCVQWLWE LYCQEAGGII 300
301 GDEMGLGKTI QIVSFLSSLH HSGKFQKPAL IVCPATLMKQ WVNEFHTWWA PLRVVVLHAT 360
361 GSGQRASREK RQYESDASES EAEESKTSIK LRGASSSFHR YAKNLVESVF TRGHILITTY 420
421 AGLRIYGDLI LPREWGYCVL DEGHKIRNPD SEISISCKQI RTVNRIILSG TPIQNNLTEL 480
481 WNLFDFVFPG RLGTLPVFQN QFALPINIGG YANASNVQVQ TAYKCACMLR DLISPYLLRR 540
541 MKLDVAADLP KKSEQVLFCK LTPLQRKAYQ DFLQGSDMQK ILNGKRQMLY GIDILRKICN 600
601 HPDLVTREYL LHKEDYNYGD PEKSGKLKVI RALLTLWKKQ GHRTLLFSQT RQMLDILEIG 660
661 LKDLPDVHYC RMDGSTSIAL RQDLVDNFNK NEYFDVFLLT TRVGGLGVNL TGADRVILFD 720
721 PDWNPSTDAQ ARERAWRLGQ KKDVVVYRLM TAGTIEEKIY HRQIFKQFLT NKILKDPKQR 780
781 RFFKMTDLHD LFTLGDNKTE GTETGSMFLG SERVLRKDNS SRNGNEAEDI PARDRKKHKI 840
841 HDKGKKVNSS KVFEKMGIAS MEKYKPPQES NVTKTNSDST LGDDSVLDDI FASAGIQSTL 900
901 KHDDIMEASQ TESILVEKEA TRVANEALRA VSSFRRPPRQ LIPPQQSTNV PGTSKPSGPI 960
961 TSSTLLARLK QRR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
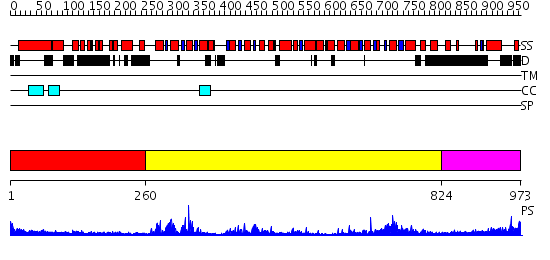
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..259] | 1.43 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain | |
| 2 | View Details | [260..823] | 87.0 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 3 | View Details | [824..973] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)