













| General Information: |
|
| Name(s) found: |
SPCC162.11c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 454 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cell division site [IDA cytoplasm [ISS spindle pole body [IDA cytosol [IDA |
| Biological Process: |
pyrimidine base metabolic process
[RCA phosphorylation [IC pyrimidine salvage [ISS |
| Molecular Function: |
ATP binding
[IEA]
uracil phosphoribosyltransferase activity [NAS] uridine kinase activity [ISS nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPEAFERYSS NPTYEPPWRK VRFIGIAGPS GSGKTSVAQL IVKALNLPHV VILSLDSFYK 60
61 SLNAEQKKRA FNNDYDFDSP EAIDWDLLFV KLLELKQGRK VDIPIYSFNE HNRLPETNTL 120
121 FGASIIILEG IFALYDEKIR SLLDVSVFLD TDSDVCLSRR LNRDINYRGR DIVGVLEQYN 180
181 KFVKPSYENF VRRQLSYTDL IVPRGRDNKL AIDMVINFIR RTLSIQSETH VKNIDSLQQI 240
241 VPTIPHLPLN LVQLKITPEI SAIRTILINK NTHPDDLQFF LSRIGTMLMN LAGDSLAYEK 300
301 KTITLHNGNQ WEGLQMAKEL CGVSVLRSGG TLETALCRQF PTVCLGKILV QINKVTQEPT 360
361 LHYHKLPRGI ATMNVVLMAS HLTTHADVLM ATQILVDFGV PEENIIIVVY VCYSESIKAL 420
421 AYIFPKVTIV TAFLESVAEP VVGRLDIEKV YYGC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
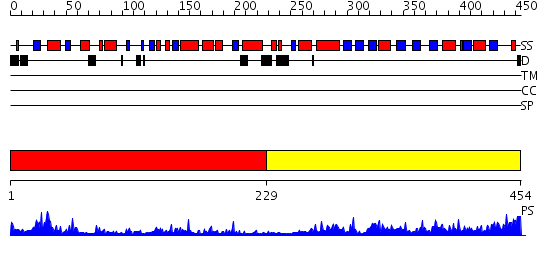
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..228] | 46.154902 | Rapid structure determination of human uridine-cytidine kinase 2 using a conventional laboratory X-ray source and a single samarium derivative | |
| 2 | View Details | [229..454] | 60.69897 | Uracil PRTase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)