













| General Information: |
|
| Name(s) found: |
rmt3 /
SPBC8D2.10c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 543 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA cytosolic small ribosomal subunit [IDA cytosol [IDA |
| Biological Process: |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine
[IPI [NOT]ribosome biogenesis [IMP ribosome assembly [IMP |
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IPI ribosome binding [IDA protein-arginine N-methyltransferase activity [IMP methyltransferase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLVKPMACYF EIWTRKVTTI EDFSLAIANR FKQMGSHSDS EVDWDNEEEV WEDEVHEFCC 60
61 LFCDSTFTCL KDLWSHCKEA HNFDFYQVKQ QNNLDFYACI KLVNYIRSQV KEGKTPDLDK 120
121 LSDILRSDEY MISVLPDDSV LFSLGDELDS DFEDDNTLEI EVENPADVSK DAEIKKLKLQ 180
181 NQLLISQLEE IRKDKMNELT SQTTDQLSVT PKKADNDSYY FESYAGNDIH FLMLNDSVRT 240
241 EGYRDFVYHN KHIFAGKTVL DVGCGTGILS MFCAKAGAKK VYAVDNSDII QMAISNAFEN 300
301 GLADQITFIR GKIEDISLPV GKVDIIISEW MGYALTFESM IDSVLVARDR FLAPSGIMAP 360
361 SETRLVLTAT TNTELLEEPI DFWSDVYGFK MNGMKDASYK GVSVQVVPQT YVNAKPVVFA 420
421 RFNMHTCKVQ DVSFTSPFSL IIDNEGPLCA FTLWFDTYFT TKRTQPIPEA IDEACGFTTG 480
481 PQGTPTHWKQ CVLLLRNRPF LQKGTRVEGT ISFSKNKKNN RDLDISVHWN VNGKADSQSY 540
541 VLN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
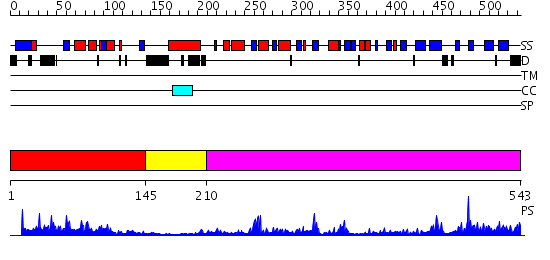
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] | 17.69897 | Solution structure of the C2H2 zinc finger domain of the protein arginine N-methyltransferase 3 from Mus musculus | |
| 2 | View Details | [145..209] | 19.0 | Crystal Structure of PH1915 (APC 5817): A Hypothetical RNA Methyltransferase | |
| 3 | View Details | [210..543] | 77.0 | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)