













| General Information: |
|
| Name(s) found: |
mst2 /
SPAC17G8.13c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 407 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IDA cytosol [IDA histone acetyltransferase complex [ISS |
| Biological Process: |
positive regulation of histone acetylation
[IMP regulation of meiotic cell cycle [IMP negative regulation of chromatin silencing at telomere [IGI [NOT]telomere maintenance [IGI histone acetylation [TAS |
| Molecular Function: |
histone acetyltransferase activity
[NAS zinc ion binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPATILKSTA SSKTLLIIKF DTNSSRYPFY KKHLLELNSE STFLGLLTKG QADTSITRLN 60
61 QDDVRLFEKA KTVADRTKDV AYSFSDPILS TQLRTPPPQP TSIRYLYFGT YRIKPWYTSP 120
121 YPEEYSCAKN LYICESCLKY MNSDHVLQRH KMKCSWSYPP GDEIYRDKNI SIFEVDGQRQ 180
181 PIYCQNLCLL AKMFLHSKML YYDVEPFLFY VLTEFDGQEC KVIGYFSKEK RSASDYNVSC 240
241 ILTLPIYQRR GYGVFLIDFS YLLTQVEGKL GSPEKPLSDL GLVTYRSYWK MRVAKALLEI 300
301 TTPISINAIA KSTSMVCDDV ISTLESLSVF KYDPLKKKYV LQLKRDELEN VYKAWNIKHP 360
361 QRVNPKLLRW TPYLGEEQIS NLLLKENILI PLPQKRLLDN SHHLDSV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
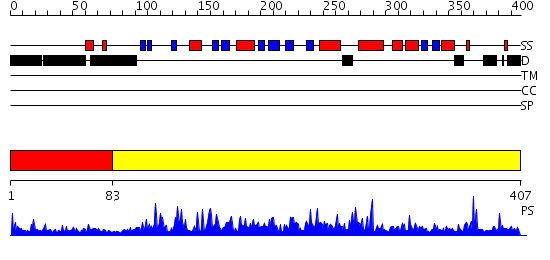
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 5.154902 | Solution Structure of the Tudor Domain from Mouse Hypothetical Protein Homologous to Histone Acetyltransferase | |
| 2 | View Details | [83..407] | 131.0 | No description for 2givA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)