













| General Information: |
|
| Name(s) found: |
pfs2 /
SPAC12G12.14c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 509 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mRNA cleavage and polyadenylation specificity factor complex [IDA cytosol [IDA [NOT]nucleolus [IDA |
| Biological Process: |
G1/S transition of mitotic cell cycle
[IMP mitotic sister chromatid segregation [IMP mRNA 3'-end processing [IMP mRNA polyadenylation [ISS mRNA cleavage [ISS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERAENARII QKPMTRRTVD YGSGLSKYIV NRHLRSNRYH IHVPRPNPNQ IINLYPPYEY 60
61 KYNNTSSLCT KYIHTSANKA RHVINVVRWT PDGRRLLTGS STGEFTLWNG LTFNFELINQ 120
121 SHDYAVRCAE WSTDGRWLIS GDGGGMVKYF EPNLNNVKIV QAHEMEVRDV AFSPNDSKFV 180
181 TASDDGSLKV WNFHMSTEEL KLTGHGWDVK TVDWHPSKGL LASGSKDNLV KFWDPRTGTC 240
241 IATLHGHKNT IMQASFQKNF GSNYLATVSR DSTCRVFDLR AMKDVRVLRG HEKDVNCVTW 300
301 HPLYPNLLTT GGSDGSVNHY SLDEPPLLSQ QKYHEKHPNV TLSASSYLLY PTAEIPFAHD 360
361 LGIWSMQYHP LGHLLCTGSN DKTTRFWSRS RPDDKESTMD RHHLGEEQSE AMLSQRKAAI 420
421 EEDDNYEPDE NPLTETLANA HNPQFSGVLN LPGLGTMPSF PSPYQHGQPQ IPGMLHASLS 480
481 NSYAEPSTQN SFIPGLTSKS QDGYPQNYR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
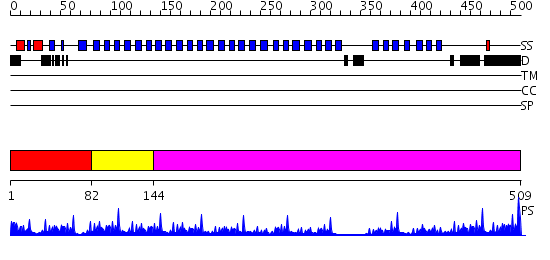
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..81] | 69.39794 | No description for 2gnqA was found. | |
| 2 | View Details | [82..143] | 24.39794 | Arp2/3 complex 41 kDa subunit ARPC1 | |
| 3 | View Details | [144..509] | 50.39794 | beta1-subunit of the signal-transducing G protein heterotrimer |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)