













| General Information: |
|
| Name(s) found: |
sid1 /
SPAC9G1.09
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 471 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spindle pole body [IDA cytosol [IDA |
| Biological Process: |
regulation of cytokinesis
[IGI septation initiation signaling [IMP protein amino acid phosphorylation [ISS |
| Molecular Function: |
ATP binding
[ISS protein binding [IPI protein serine/threonine kinase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYPLNANSYT LLRKLGSGSF GVVWKARENV SGDIIAIKQI DLETGIDDIT DIEQEVFMLS 60
61 NCNSSNVIQY YGCFVDGYTL WILMEHMDGG SVSGLLKMGR LNEQVISIIL REVLYGLNYL 120
121 HGQNKIHRDI KAANILLSSS TGNVKLADFG VAAQLSNAAS RRHTFVGTPF WMAPEVIQQT 180
181 SYGLAADIWS LGITAIEMAN GIPPRATMHP MRVIFEIPQS EPPKLDDHFS PTFRDFVSCC 240
241 LDLNPNMRWS AKELLQHPFI KSAGTVKDII PLLVQKENKL FDDSDQSVLE ETINNTLKPF 300
301 EEPIAEGNAD IEDWTFETVK KSDSTVLGNT SIPKNSIISS QNKEELPSSI KYLEKTIMSD 360
361 QATPHPFSKS LSEKGSSYHK SLTSDFAMKH YIKSTIRSML LNDKLSATQR SSLESFYTSF 420
421 ISLDKNLSSK FVNQITPDNR LHHKKQKRSP ISQLLFSRWL EETEKRRSLN G |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
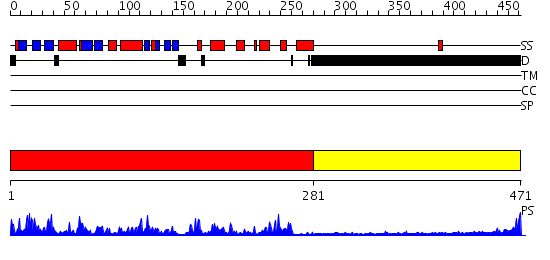
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..280] | 63.69897 | No description for 2j51A was found. | |
| 2 | View Details | [281..471] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)