













| General Information: |
|
| Name(s) found: |
shk2 /
SPAC1F5.09c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 589 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
MAPKKK cascade
[IGI protein amino acid phosphorylation [ISS regulation of conjugation with cellular fusion by signal transduction [NAS] |
| Molecular Function: |
ATP binding
[ISS protein binding [IPI protein serine/threonine kinase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLLSVRGVPV EIPSLKDTKK SKGIIRSGWV MLKEDKMKYL PWTKKWLVLS SNSLSIYKGS 60
61 KSESAQVTLL LKDIQKVERS KSRTFCFKLR FKSSTKNFEI QACELSVADN MECYEWMDLI 120
121 SSRALASKVS SPMNPKHQVH VGIDNEGNYV GLPKEWILLL QSSSITKQEC MEEPKAVIQA 180
181 LDFYSKQLDT TSETKDSFSF CKETLPRSST TSYSIRDADK HHKLTTSGVT KMNITERCKP 240
241 KTTIQTDKKH IIRPFIEDKS HVESIMTGKV TKVPVKADSK NTLSRRMTDR QALAMLKDSV 300
301 TSHDPVEYFN VKHKLGQGAS GSVYLAKVVG GKQLGIFDSV AIKSIDLQCQ TRKELILNEI 360
361 TVMRESIHPN IVTYLDSFLV RERHLWVVME YMNAGSLTDI IEKSKLTEAQ IARICLETCK 420
421 GIQHLHARNI IHRDIKSDNV LLDNSGNIKI TDFGFCARLS NRTNKRVTMV GTPYWMAPEV 480
481 VKQNEYGTKV DIWSLGIMII EMIENEPPYL REDPIRALYL IAKNGTPTLK KPNLVSKNLK 540
541 SFLNSCLTID TIFRATAAEL LTHSFLNQAC PTEDLKSIIF SRKANTHIN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
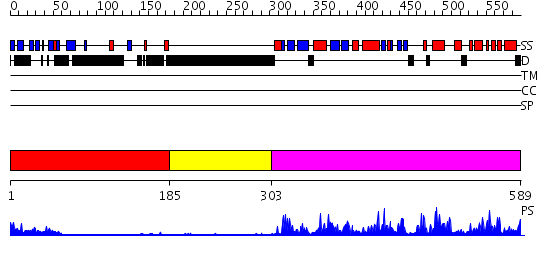
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | 3.69897 | Fibroblast growth factor receptor, FGFR | |
| 2 | View Details | [185..302] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [303..589] | 78.0 | pak1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)