













| General Information: |
|
| Name(s) found: |
rfc1 /
SPBC23E6.07c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 934 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Ctf18 RFC-like complex
[TAS Elg1 RFC-like complex [IDA nucleus [IDA nuclear chromatin [IC nuclear replication fork [IC] nucleolus [IEA] DNA replication factor C complex [TAS Rad17 RFC-like complex [TAS |
| Biological Process: |
DNA repair
[ISS DNA-dependent DNA replication [IGI leading strand elongation [ISS |
| Molecular Function: |
ATP binding
[IEA]
DNA clamp loader activity [IC protein binding [IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNSDIRSFF GGGNAQKKPK VSPTPTSPKP KRSLKKKRIV LSDDEDGTIE NSKVPASKSK 60
61 VQKRNESEDI SHSLPSIVHE DDKLVGSDGV STTPDEYFEQ QSTRSRSKPR IISNKETTTS 120
121 KDVVHPVKTE NFANDLDTTS DSKPVVHQTR ATRKPAQPKA EKSTTSKSKS HTTTATTHTS 180
181 RSSKSKGLPR FSDEVSQALK NVPLIDVDSM GVMAPGTFYE RAATTQTPGS KPVPEGNSDC 240
241 LSGISFVITG ILETLTRQEA TDLIKQYGGK VTGAPSVRTD FILLGENAGP RKVETIKQHK 300
301 IPAINEDGLF YLITHLPASG GTGAAAQAAQ QKKEQEEKKI LETVARMDDS NKKESQPSQI 360
361 WTSKYAPTSL KDICGNKGVV QKLQKWLQDY HKNRKSNFNK PGPDGLGLYK AVLLSGPPGI 420
421 GKTTAAHLVA KLEGYDVLEL NASDTRSKRL LDEQLFGVTD SQSLAGYFGT KANPVDMAKS 480
481 RLVLIMDEID GMSSGDRGGV GQLNMIIKKS MIPIICICND RAHPKLRPLD RTTFDLRFRR 540
541 PDANSMRSRI MSIAYREGLK LSPQAVDQLV QGTQSDMRQI INLLSTYKLS CSEMTPQNSQ 600
601 AVIKNSEKHI VMKPWDICSR YLHGGMFHPS SKSTINDKLE LYFNDHEFSY LMVQENYLNT 660
661 TPDRIRQEPP KMSHLKHLEL ISSAANSFSD SDLVDSMIHG PQQHWSLMPT HALMSCVRPA 720
721 SFVAGSGSRQ IRFTNWLGNN SKTNKLYRML REIQVHMRLK VSANKLDLRQ HYIPILYESL 780
781 PVKLSTGHSD VVPEIIELMD EYYLNREDFD SITELVLPAD AGEKLMKTIP TAAKSAFTRK 840
841 YNSSSHPIAF FGSSDVLPMK GSAQREVPDV EDAIEAEDEM LEEASDSEAA NEEDIDLSKD 900
901 KFISVPKKPK KRTKAKAEAS SSSSTSRRSR KKTA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
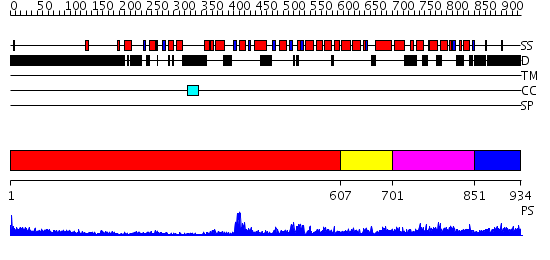
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..606] | 20.0 | N-terminal, ClpS-binding domain of ClpA, an Hsp100 chaperone; ClpA, an Hsp100 chaperone, AAA+ modules | |
| 2 | View Details | [607..700] | 41.69897 | Replication factor C | |
| 3 | View Details | [701..850] | 60.0 | Crystal Structure of the Eukaryotic Clamp Loader (Replication Factor C, RFC) Bound to the DNA Sliding Clamp (Proliferating Cell Nuclear Antigen, PCNA) | |
| 4 | View Details | [851..934] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)