













| General Information: |
|
| Name(s) found: |
ssb1 /
SPBC660.13c
[Sanger Pombe]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 609 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromosome, telomeric region
[ISS nucleus [IDA cytoplasm [IDA cytosol [IDA DNA replication factor A complex [IDA |
| Biological Process: |
DNA repair
[IMP DNA-dependent DNA replication [IMP double-strand break repair via homologous recombination [ISS telomere maintenance [IGI DNA replication, synthesis of RNA primer [ISS |
| Molecular Function: |
zinc ion binding
[IEA]
single-stranded DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAERLSVGAL RIINTSDASS FPPNPILQVL TVKELNSNPT SGAPKRYRVV LSDSINYAQS 60
61 MLSTQLNHLV AENKLQKGAF VQLTQFTVNV MKERKILIVL GLNVLTELGV MDKIGNPAGL 120
121 ETVDALRQQQ NEQNNASAPR TGISTSTNSF YGNNAAATAP APPPMMKKPA APNSLSTIIY 180
181 PIEGLSPYQN KWTIRARVTN KSEVKHWHNQ RGEGKLFSVN LLDESGEIRA TGFNDQVDAF 240
241 YDILQEGSVY YISRCRVNIA KKQYTNVQNE YELMFERDTE IRKAEDQTAV PVAKFSFVSL 300
301 QEVGDVAKDA VIDVIGVLQN VGPVQQITSR ATSRGFDKRD ITIVDQTGYE MRVTLWGKTA 360
361 IEFSVSEESI LAFKGVKVND FQGRSLSMLT SSTMSVDPDI QESHLLKGWY DGQGRGQEFA 420
421 KHSVISSTLS TTGRSAERKN IAEVQAEHLG MSETPDYFSL KGTIVYIRKK NVSYPACPAA 480
481 DCNKKVFDQG GSWRCEKCNK EYDAPQYRYI ITIAVGDHTG QLWLNVFDDV GKLIMHKTAD 540
541 ELNDLQENDE NAFMNCMAEA CYMPYIFQCR AKQDNFKGEM RVRYTVMSIN QMDWKEESKR 600
601 LINFIESAQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
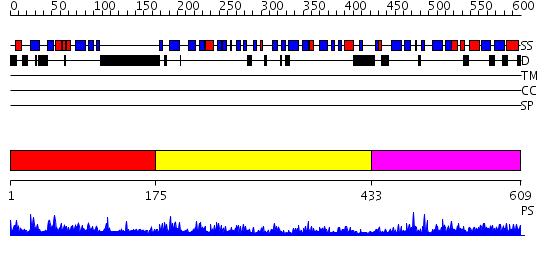
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..174] | 35.522879 | N-terminal domain of the RPA70 subunit of human replication protein A. | |
| 2 | View Details | [175..432] | 67.30103 | Replication protein A 70 KDa subunit (RPA70) | |
| 3 | View Details | [433..609] | 51.39794 | Replication protein A 70 KDa subunit (RPA70) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)