













| General Information: |
|
| Name(s) found: |
rhp55 /
SPAC3C7.03c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 350 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Rhp55-Rhp57 complex [NAS] cytosol [IDA |
| Biological Process: |
meiotic joint molecule formation
[TAS reciprocal meiotic recombination [IMP recombinational repair [IMP double-strand break repair via homologous recombination [IGI meiotic gene conversion [IMP response to DNA damage stimulus [IMP DNA recombinase assembly [NAS] nucleus organization [IMP strand invasion [TAS |
| Molecular Function: |
ATP binding
[ISS DNA binding [IEA] DNA-dependent ATPase activity [ISS protein-DNA loading ATPase activity [TAS protein heterodimerization activity [IPI single-stranded DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSSQHRLVT QPAIRAYEAF SAPGFGFNSK LLDDAFGGSG LKRGYISEVC GAPGMGKTSL 60
61 ALQITANALL SGSRVIWVET CQPIPMERLR QLLDNHVPSS QDEEEKCDTD ELLNLLDVVY 120
121 APNLVNILAF LRNFDQEKHL KEIGLLIIDN LSMPIQLAYP TSPEDYAYLR LRRNTSKKSS 180
181 LSDSSQKENT LTLNKENEFS SKDDSNFAFH NSSTKTTINR RKKAIGTISS LLSKITSSCY 240
241 VAIFVTTQMT SKVVSGIGAK LIPLLSTNWL DNLSYRLILY SRHSTEESKD GQSRPSHQLL 300
301 RYAFMAKQPP AHSAESELAF QLTSTGIQDY QSIPTNSSQR RKRSILECES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
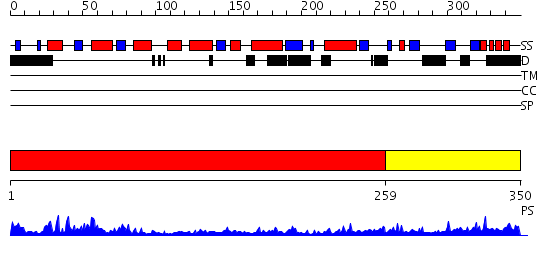
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..258] | 50.39794 | RecA | |
| 2 | View Details | [259..350] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)