













| General Information: |
|
| Name(s) found: |
exo1 /
SPBC29A10.05
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 571 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spindle [IDA |
| Biological Process: |
maintenance of DNA repeat elements
[IMP meiotic DNA double-strand break processing [TAS telomere maintenance [TAS mismatch repair [IGI mitotic recombination [TAS |
| Molecular Function: |
5'-3' exonuclease activity
[IMP magnesium ion binding [IEA] 5'-flap endonuclease activity [ISS double-stranded DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGIKGLLGLL KPMQKSSHVE EFSGKTLGVD GYVWLHKAVF TCAHELAFNK ETDKYLKYAI 60
61 HQALMLQYYG VKPLIVFDGG PLPCKASTEQ KRKERRQEAF ELGKKLWDEG KKSQAIMQFS 120
121 RCVDVTPEMA WKLIIALREH GIESIVAPYE ADAQLVYLEK ENIIDGIITE DSDMLVFGAQ 180
181 TVLFKMDGFG NCITIRRNDI ANAQDLNLRL PIEKLRHMAI FSGCDYTDGV AGMGLKTALR 240
241 YLQKYPEPRA AIRAMRLDKS LKVPVSFEKE FALADLAFRH QRVYCPKDKT LVHLSPPERE 300
301 LSVHEDAFIG SFFDNQLAID IAEGRSNPIT KCAFDIKDSS MQSFTKTTIT ISKRKGISKT 360
361 DISNFFMKSI PPSKRPTKST SLIDVTNVKV QRTHLANDIS SEKQSIKSAN EKAYVTPKSN 420
421 SLKPGFGKSL SDISNSATKN ENVPFLPPRT GVSKYFKLQK NTEKEIDEQV PSQSNNTTPT 480
481 SAKSDSASPQ NWFSSFSYQT PNSASPPFSS LSHTLPISAL AKIGHDALNR KNHASLPSRR 540
541 IVYKPPSSPS TPISMNPRPK GILSLQQYKF R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
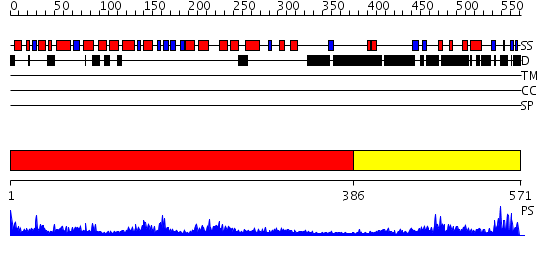
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..385] | 73.69897 | Crystal structure of the human FEN1-PCNA complex | |
| 2 | View Details | [386..571] | 45.0 | STRUCTURE OF LARGE FRAGMENT OF ESCHERICHIA COLI DNA POLYMERASE I COMPLEXED WITH D/TMP |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)