













| General Information: |
|
| Name(s) found: |
nth1 /
SPAC30D11.07
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 355 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrial matrix [ISS |
| Biological Process: |
base-excision repair
[IMP base-excision repair, AP site formation [IDA |
| Molecular Function: |
5-formyluracil DNA N-glycosylase activity
[IDA DNA binding [ISS iron ion binding [IEA] endonuclease activity [IEA] 5-hydroxymethyluracil DNA N-glycosylase activity [IDA oxidized pyrimidine base lesion DNA N-glycosylase activity [IMP 8-oxo-7,8-dihydroguanine DNA N-glycosylase activity [IDA 4 iron, 4 sulfur cluster binding [IEA] iron-sulfur cluster binding [IEA] oxidized purine base lesion DNA N-glycosylase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKDYGTPPE NWREVYDEIC KMKAKVVAPV DVQGCHTLGE RNDPKKFRFQ TLVALMLSSQ 60
61 TKDIVLGPTM RNLKEKLAGG LCLEDIQNID EVSLNKLIEK VGFHNRKTIY LKQMARILSE 120
121 KFQGDIPDTV EDLMTLPGVG PKMGYLCMSI AWNKTVGIGV DVHVHRICNL LHWCNTKTEE 180
181 QTRAALQSWL PKELWFELNH TLVGFGQTIC LPRGRRCDMC TLSSKGLCPS AFKEKSGITI 240
241 TKRKVKTIKR VKKRPASESP PLSPLSLPTD DLYYQSIEDK SLIKLEDLDP VDSISHMNEP 300
301 LKKEPAADID VDQKPPVAFH STTKETRSLR RSKRVAKKSS QYFSQQSLQD IEDLV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
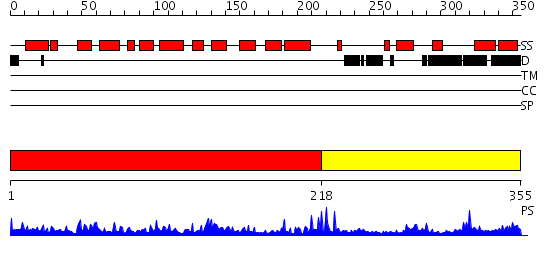
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..217] | 56.221849 | REFINEMENT OF THE NATIVE STRUCTURE OF ENDONUCLEASE III TO A RESOLUTION OF 1.85 ANGSTROM | |
| 2 | View Details | [218..355] | 6.69897 | Solution structure of the NUDIX domain from human A/G-specific adenine DNA glycosylase alpha-3 splice isoform |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)