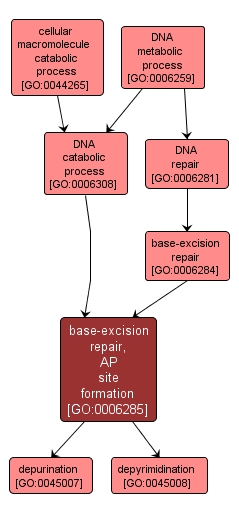
GO TERM SUMMARY
|
| Name: |
base-excision repair, AP site formation |
| Acc: |
GO:0006285 |
| Aspect: |
Biological Process |
| Desc: |
The formation of an AP site, a deoxyribose sugar with a missing base, by DNA glycosylase which recognizes an altered base in DNA and catalyzes its hydrolytic removal. This sugar phosphate is the substrate recognized by the AP endonuclease, which cuts the DNA phosphodiester backbone at the 5' side of the altered site to leave a gap which is subsequently repaired. |
|

|
INTERACTIVE GO GRAPH
|