













| General Information: |
|
| Name(s) found: |
cwf24 /
SPBC13E7.02
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 533 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spliceosomal complex [IDA |
| Biological Process: |
snoRNA metabolic process
[NAS]
nuclear mRNA splicing, via spliceosome [TAS protein ubiquitination [ISS |
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS DNA binding [IEA] zinc ion binding [ISS N-acetyltransferase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQKNLNINQ ASGSKINTEL AVPIFQSRRR HRPRQGLKRK KGFKRDDDSG GSSESSNEDM 60
61 RDNIPIVSGR KKTVRLNRLQ RESEQFENSA LKDINVEYQS NLSATGESVN TTTVSAINED 120
121 TREVILGRPS PKLANQSTLP TELFQSQNDY SRFLPKRKDF EKKSQVGPVL SSNASTVRMN 180
181 TIIDYQPDVC KDYKLTGYCG YGDTCKFLHM REDYKAGWQL DREWDSVQEK YKKGAKLEEG 240
241 MVKNEKKEDI PFVCLICKKD YRSPIATTCG HHFCEQCAIT RYRKTPTCIQ CGADTKGLFS 300
301 VDKNFDRLLK NRKSKNDEAV KQKVGGFESN NSATTEVSER KDREASFQGF ADTLAKPNTS 360
361 AQQKMPSLGD NSNTIISKYF IREITESNIV HFKRLVRVVL EASYSDKFYR LVLKNPDYAR 420
421 IATFEDKFVG AISSLVAEDN SLYVTVLCVL APYRCLGIGS LLIDHVKKTA INNNIDRISL 480
481 HVQTTNESVI KWYTAHGFKI VKQINDFYRR LENKSAFYMV CPLSAHNNII SNH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
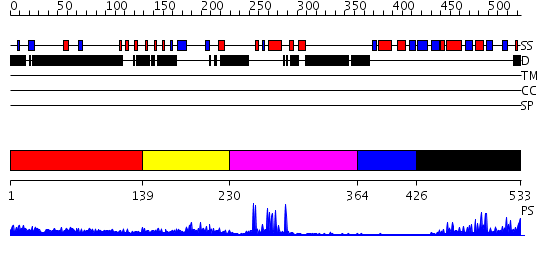
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [139..229] | 1.49 | Solution Structure of the Zinc-finger domain in KIAA1064 protein | |
| 3 | View Details | [230..363] | 13.09691 | RAG1 DIMERIZATION DOMAIN | |
| 4 | View Details | [364..425] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [426..533] | 15.221849 | No description for 2ob0A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)