













| General Information: |
|
| Name(s) found: |
alp13 /
SPAC23H4.12
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 337 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA NuA4 histone acetyltransferase complex [IDA nuclear chromatin [IC Rpd3S complex [IDA |
| Biological Process: |
chromatin remodeling
[IC chromosome condensation [IMP establishment or maintenance of cell polarity [IMP chromatin assembly or disassembly [IEA] histone acetylation [IC] histone deacetylation [IGI response to DNA damage stimulus [IMP regulation of transcription from RNA polymerase II promoter [ISS |
| Molecular Function: |
protein binding
[IPI chromatin binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVSYKVNER VLCFHGPLLY EAKIVDTEMK GDVTTYLIHY KGWKNSWDEW VEQDRILQWT 60
61 EENLKTQKEL KNAAISTRQK PTSKKSASST SKHDSTGVKT SGKRSRESST VTVDGDSHEL 120
121 PSRIKTQKSE SPIPQQVKRD GTTDAKNEET TKPENNEKDD FEEEPPLPKH KISVPDVLKL 180
181 WLVDDWENIT KNQQLIAIPR NPTVRAAIAA FRESKISHLN NEIDVDVFEQ AMAGLVIYFN 240
241 KCLGNMLLYR FERQQYLEIR QQYPDTEMCD LYGVEHLIRL FVSLPELIDR TNMDSQSIEC 300
301 LLNYIEEFLK YLVLHKDEYF IKEYQNAPPN YRSLVGV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
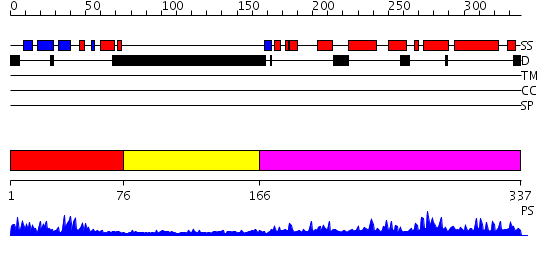
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | 23.0 | No description for 2efiA was found. | |
| 2 | View Details | [76..165] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [166..337] | 58.69897 | Crystal Structure of the MRG15 MRG domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)