













| General Information: |
|
| Name(s) found: |
cbp1 /
SPBC1105.04c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 522 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA condensed nuclear chromosome, centromeric region [IDA nuclear chromatin [IC |
| Biological Process: |
DNA replication initiation
[IGI chromatin silencing at centromere [IMP histone modification [IMP chromosome segregation [IGI |
| Molecular Function: |
protein binding
[IPI protein dimerization activity [NAS centromeric DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKIKRRAIT EHEKRALRHY FFQLQNRSGQ QDLIEWFREK FGKDISQPSV SQILSSKYSY 60
61 LDNTVEKPWD VKRNRPPKYP LLEAALFEWQ VQQGDDATLS GETIKRAAAI LWHKIPEYQD 120
121 QPVPNFSNGW LEGFRKRHIL HAINEQPTES VVLNNTEPPN DPLSRVYDVT RLTNINDIFT 180
181 MQETGLFWKL VPNGTPEVED IKGITRFKAR ITLTVCCNAS GTERLPLWVI GYSQSPRVFR 240
241 AANVKPEVMN FKWRSNGKAS MTTAIMEEWL RWFDACMEGR KVILLIDSYT PHLRAVENIR 300
301 NSGNDLRNTT VITLPSTSAS ISQPCSEGVI YALKACYRKH WVQYILEQNE LGRNPYNTTN 360
361 VLRAILWLVK AWTTDISPEI IENAFNLSGV LGLFNESAVT SRALDEMIHP LRELVSEFSV 420
421 QAAMRIEDFI SPSEENIVDS SEDIINQIAS QYMDDRAFET DEEESTEFQI TTKDAMKAIE 480
481 LLLNYEAQQP DGNPAITISL LNYQKLLEAR GGNVNLSRLR ST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
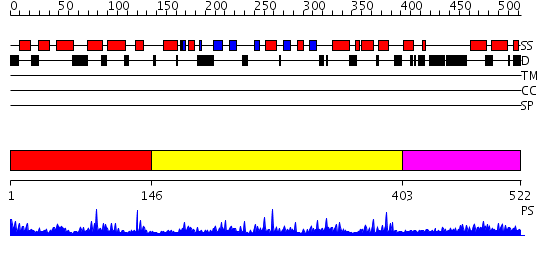
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..145] | 29.154902 | Ars-binding protein 1, ABP1 | |
| 2 | View Details | [146..402] | 1.34 | No description for 2f7tA was found. | |
| 3 | View Details | [403..522] | 3.146991 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)