













| General Information: |
|
| Name(s) found: |
pcm-PA /
FBpp0074553
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1612 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[NAS]
cytoplasmic mRNA processing body [IDA] |
| Biological Process: |
dorsal closure
[IMP]
wound healing [IMP] imaginal disc fusion, thorax closure [IMP] nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [IMP] spermatogenesis [IMP] imaginal disc-derived wing morphogenesis [IMP] nuclear-transcribed mRNA catabolic process, exonucleolytic, 5'-3' [IMP] nucleobase, nucleoside, nucleotide and nucleic acid metabolic process [IEA] RNA interference [IMP] RNA catabolic process [NAS] gastrulation [NAS] |
| Molecular Function: |
nucleic acid binding
[IEA]
5'-3' exoribonuclease activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVPKFFRYI SERYPCLSEL AREHCIPEFD NLYLDMNGIV HNCSHPDDNN IHFHLEEEQI 60
61 FQEIFNYVDK LFYLIKPQRL FFLSVDGVAP RAKMNQQRSR RFRTAREAEQ QEAKAAQRGE 120
121 LREHERFDSN CITPGTEFMV RLQEGLRAFL KTKISTDPLW QRCTVILSGQ EAPGEGEHKI 180
181 MDYIRYMKTQ PDYDPNTRHC LYGLDADLII LGLCTHELHF VVLREEVKFG RNVKRTSVEE 240
241 TRFFLLHLGL LREYLELEFD ALRTDEHKLD IAQLIDDWVL MGFLVGNDFI PHLPCLHISS 300
301 NALPLLYRTY IGIYPTLGGN INENGKLNLR RLQIFISALT EVELDHFKEH ADDLKYMNNK 360
361 SEAFDMDVGE ITESQNLDSD LGALINKSML LYDDDSEEDC SDENAVLLKE FQNYKRNFYR 420
421 NKFKRDPNDE LIEELCHHYV NALQWVLDYY YRGVQSWDWY YPFHYTPFIS DLKNIEQVEI 480
481 AFHMGTPFLP FQQLLAVLPA ASAKLLPVAY HDLMLLPTSP LAEFYPLEFE SDLNGKKHDW 540
541 EAVVLIPFID EGRLLAAMLP CEAQLSLEER ERNRHGPMYV YKYSTVAQGP MPAYPPLRAL 600
601 PVLYCTEVAK WSHEIAVNLP YSVCIELPNA ARTVFFPGFP TMQHLPFDFE LRNDRVKVFE 660
661 QVSRNQNIVL KPRKRQLEDT LTAVASQYLG KVIHVGWPHL VKAIVVRVAT RDQRVDSEGI 720
721 TLNDSRRFDS ECKALQEHFI NRMGIQFANY DVLVYVRTFA GNSTEFRDKG ALMVRDSWSS 780
781 SVTGYPAQGV VADLTVWERM RKNFLNVEHY FPVGSTIFLI TDPYYGSEGT VQDPRLAYTN 840
841 GRIQVSIMVR PEPKVNAARQ LQEERDRDYL STFQVCNLLR ISGRTLGRLS GTVWVVLGPR 900
901 RQKMENVTKH NIGLQLKYPR QNEERAGYCF RTNNQWYYSS LAVDLMRNYC QRYPDVIDFF 960
961 GDSNDRAEFV FEQDVFPNAV GHRRVEELAN WVRQQPHMKV ERISCGSKTV CRETIELLIA1020
1021 AVDDLRSLPV KHVKLQVKPH LLIKPNVTLP DVYRSKRPVR LFDRVVIVRT IYMVPVGTKG1080
1081 TVIGIHPVTD PNPVRLECVH AVDTFCKVLF DSPVPNCNNI HGIAEDRVYK VPEIALVIIK1140
1141 TDEEGKKQND CELPVRDPQP NQAQDEPVRA TSSRYVTAAG STSVPITMKT QISDEFVKTR1200
1201 SDPIARTDSY KPSSEPKPVP VPEQITNWRE RVSTPTNKPQ PAPNNWRINR SSSRQQGGSI1260
1261 FVAPPTKTPD AAASTASTAF TAASSATLTP LDQTLALMSV LGVGEDQSSP PLQEAVQQQR1320
1321 PPLLQQQRAP FPGQMPNLPK PPLFWQQEAQ KQEALQQEAQ QQEAQKKQQQ AHAQMEPERI1380
1381 NSQHFYRSGQ TGAALNQPPL GAPSKRQWHE WVHPRMQHAN AFHAGVNNGY QMRPKKNIAA1440
1441 QSTFNNNVHM HLQQPYYPNQ QQQQQQQQPL QLTEINNAPP RYSTIQDFVP IQAYRPKKLN1500
1501 RVQPAGRQDV DATKNPSRSP VLQQPTNETI DTKASSSLPV QSAGEQVIGL MQTLEIKPAA1560
1561 SQSESDGVST GSANAPTATT SSQAVNRRKH RVPRIGAKFD LEYILPDSPH PT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
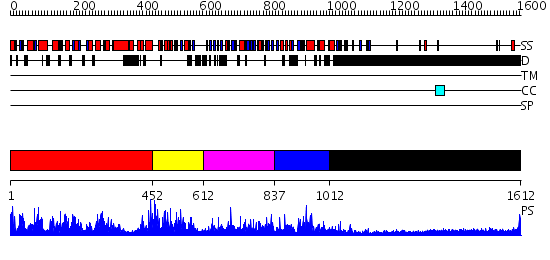
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..451] | 1.85 | Crystal structure of the human FEN1-PCNA complex | |
| 2 | View Details | [452..611] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [612..836] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [837..1011] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1012..1612] | 8.39794 | No description for 1y0fA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)