













| General Information: |
|
| Name(s) found: |
psh-PA /
FBpp0074400
[FlyBase]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 394 amino acids |
Gene Ontology: |
|
| Cellular Component: |
extracellular region
[IDA]
|
| Biological Process: |
positive regulation of antifungal peptide biosynthetic process
[IMP]
innate immune response [IDA] proteolysis [IMP][NAS][TAS] regulation of Toll signaling pathway [TAS] defense response to fungus [IMP][TAS] |
| Molecular Function: |
serine-type endopeptidase activity
[IEA][NAS]
peptidase activity [IMP][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLKWSLLLG TFVLISCSSV EAAVTVGRAC KVTDTMPGIC RTSSDCEPLI DGYIKSGVLT 60
61 LNDVPSCGLG AWGEIFCCPT KPCCDNSTIT SVSTSSTTST KAPMTSGRVD VPTFGSGDRP 120
121 AVAACKKIRE RKQQRSGNQL VIHIVGGYPV DPGVYPHMAA IGYITFGTDF RCGGSLIASR 180
181 FVLTAAHCVN TDANTPAFVR LGAVNIENPD HSYQDIVIRS VKIHPQYVGN KYNDIAILEL 240
241 ERDVVETDNI RPACLHTDAT DPPSNSKFFV AGWGVLNVTT RARSKILLRA GLELVPLDQC 300
301 NISYAEQPGS IRLLKQGVID SLLCAIDQKL IADACKGDSG GPLIHELNVE DGMYTIMGVI 360
361 SSGFGCATVT PGLYTRVSSY LDFIEGIVWP DNRV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
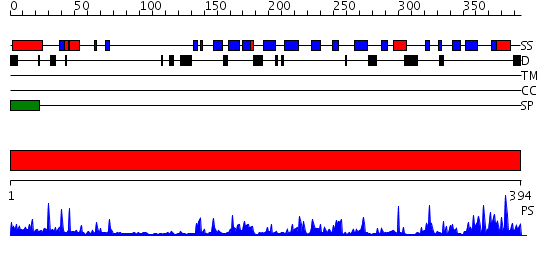
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..394] | 58.0 | Crystal structure of the extracellular region of the transmembrane serine protease hepsin with covalently bound preferred substrate. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|