













| General Information: |
|
| Name(s) found: |
HDAC6-PA /
FBpp0073825
[FlyBase]
|
| Description(s) found:
Found 37 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1128 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
|
| Biological Process: |
histone deacetylation
[IDA]
regulation of transcription [IDA] |
| Molecular Function: |
zinc ion binding
[IEA]
histone deacetylase activity [IDA][ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSPPIVTRRS AQQAKIQTRA MASKSKTGTT GAATAAGGTG SSSGSISGGF NAADPRKSNK 60
61 PNAALLEAKR RARNNMLKNQ GCGSQESVTD IFQNAVNSKS LVRKPTALIY DESMSQHCCL 120
121 WDKEHYECPE RFTRVLERCR ELNLTERCLE LPSRSATKDE ILRLHTEEHF ERLKETSGIR 180
181 DDERMEELSS RYDSIYIHPS TFELSLLASG STIELVDHLV AGKAQNGMAI IRPPGHHAMK 240
241 AEYNGYCFFN NVALATQHAL DVHKLQRILI IDYDVHHGQG TQRFFYNDPR VVYFSIHRFE 300
301 HGSFWPHLHE SDYHAIGSGA GTGYNFNVPL NATGMTNGDY LAIFQQLLLP VALEFQPELI 360
361 IVSAGYDAAL GCPEGEMEVT PACYPHLLNP LLRLADARVA VVLEGGYCLD SLAEGAALTL 420
421 RSLLGDPCPP LVETVPLPRA ELAQALLSCI AVHRPHWRCL QLQQTYDCVE LQDRDKEEDL 480
481 HTVLRHWIGG PPPMDRYPTR DTAIPLPPEK LTSNAARLQV LRAETKLSVP SFKVCYAYDA 540
541 QMLLHCNLND TGHPEQPSRI QHIHKMHDDY GLLKQMKQLS PRAATTDEVC LAHTRAHVNT 600
601 VRRLLGREPK ELHDAAGIYN SVYLHPRTFD CATLAAGLVL QAVDSVLRGE SRSGICNVRP 660
661 PGHHAEQDHP HGFCIFNNVA IAAQYAIRDF GLERVLIVDW DVHHGNGTQH IFESNPKVLY 720
721 ISLHRYEHGS FFPKGPDGNF DVVGKGAGRG FNVNIPWNKK GMGDLEYALA FQQLIMPIAY 780
781 EFNPQLVLVS AGFDAAIGDP LGGCKVTAEG YGMLTHWLSA LASGRIIVCL EGGYNVNSIS 840
841 YAMTMCTKTL LGDPVPTPQL GATALQKPPT VAFQSCVESL QQCLQVQRNH WRSLEFVGRR 900
901 LPRDPVVGEN NNEDFLTASL RHLNISNDDA TAAAGGLAGD RPDCGDERPS GSKPKVKVKT 960
961 LSDYLAENKE ALEQSEMFAV YPLKTCPHLR LLRPEEAPRS LDSGAECSVC GSTGENWVCL1020
1021 SCRHVACGRY VNAHMEQHSV EEQHPLAMST ADLSVWCYAC SAYVDHPRLY AYLNPLHEDK1080
1081 FQEPMAWTHG CAWREDGCYA TGPDGRDEDD DDDNGAGSSI CLRLERNN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
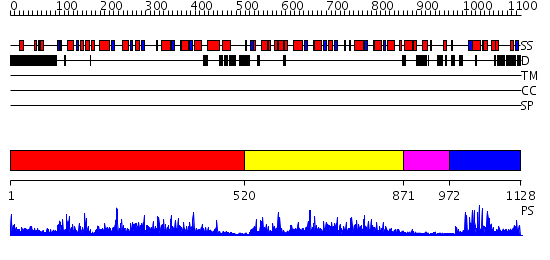
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..519] | 108.0 | No description for 2nvrA was found. | |
| 2 | View Details | [520..870] | 66.0 | HDAC homologue | |
| 3 | View Details | [871..971] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [972..1128] | 24.39794 | No description for 2g43A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|