













| General Information: |
|
| Name(s) found: |
FBpp0309145
[FlyBase]
Smox-PA / FBpp0071140 [FlyBase] |
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 486 amino acids |
Gene Ontology: |
|
| Cellular Component: |
transcription factor complex
[IEA]
|
| Biological Process: |
mushroom body development
[IMP]
imaginal disc-derived wing morphogenesis [IMP] transforming growth factor beta receptor signaling pathway [IEA] regulation of transcription, DNA-dependent [IEA] axon guidance [IMP] neuron development [IMP] regulation of mitotic cell cycle [IMP] dendrite morphogenesis [NAS] |
| Molecular Function: |
transcription factor activity
[IEA]
protein binding [IEA] transforming growth factor beta receptor, cytoplasmic mediator activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLPFTPQVVK RLLALKKGNE DNSVEGKWSE KAVKNLVKKI KKNSQLEELE RAISTQNCQT 60
61 RCVTVPRSKP APAGEHLRKG LPHVIYCRLW RWPDLQSQNE LKPLDHCEYA FHLRKEEICI 120
121 NPYHYKKIEL SILVPKSLPT PPDSIVDYPL DNHTHQIPNN TDYNAAIIRS ASLSPPQYME 180
181 LGGAGPVSVS SSASSTPATA AGGGGGPSSS SSSSSSAASA YQQQQQQLSF GQNMDSQSSV 240
241 LSVGSSIPNT GTPPPGYMSE DGDPIDPNDN MNMSRLTPPA DAAPVMYHEP AFWCSISYYE 300
301 LNTRVGETFH ASQPSITVDG FTDPSNSERF CLGLLSNVNR NEVVEQTRRH IGKGVRLYYI 360
361 GGEVFAECLS DSSIFVQSPN CNQRYGWHPA TVCKIPPGCN LKIFNNQEFA ALLSQSVSQG 420
421 FEAVYQLTRM CTIRMSFVKG WGAEYRRQTV TSTPCWIELH LNGPLQWLDR VLTQMGSPRL 480
481 PCSSMS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
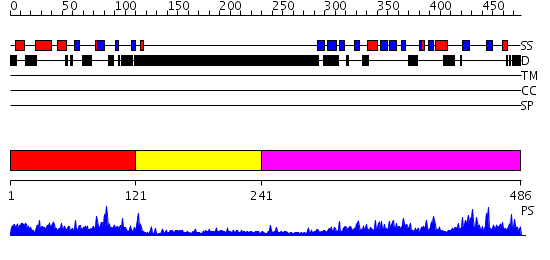
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 54.30103 | SMAD MH1 domain | |
| 2 | View Details | [121..240] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [241..486] | 94.39794 | Smad2 MH2 domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)