













| General Information: |
|
| Name(s) found: |
FBpp0309041
[FlyBase]
Spt6-PA / FBpp0070861 [FlyBase] |
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1831 amino acids |
Gene Ontology: |
|
| Cellular Component: |
polytene chromosome
[IDA]
nuclear exosome (RNase complex) [IPI] polytene chromosome puff [IDA] polytene chromosome interband [IDA] |
| Biological Process: |
RNA elongation
[NAS]
transcription from RNA polymerase II promoter [IEP] nucleobase, nucleoside, nucleotide and nucleic acid metabolic process [IEA] regulation of transcription from RNA polymerase II promoter [IEA] |
| Molecular Function: |
hydrolase activity, acting on ester bonds
[IEA]
transcription elongation regulator activity [IEA][NAS] protein binding [IEA] RNA binding [IEA] chromatin binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEFLDSEAE ESEEEEELDV NERKRLKKLK AAVSDSSEEE EDDEERLREE LKDLIDDNPI 60
61 EEDDGSGYDS DGVGSGKKRK KHEDDDLDDR LEDDDYDLIE ENLGVKVERR KRFKRLRRIH 120
121 DNESDGEEQH VDEGLVREQI AEQLFDENDE SIGHRSERSH READDYDDVD TESDADDFIV 180
181 DDNGRPIAEK KKKRRPIFTD ASLQEGQDIF GVDFDYDDFS KYEEDDYEDD SEGDEYDEDL 240
241 GVGDDTRVKK KKALKKKVVK KTIFDIYEPS ELKRGHFTDM DNEIRKTDIP ERMQLREVPV 300
301 TPVPEGSDEL DLEAEWIYKY AFCKHTVSEQ EKPESREKMR KPPTTVNKIK QTLEFIRNQQ 360
361 LEVPFIAFYR KEYVKPELNI DDLWKVYYYD GIWCQLNERK RKLKVLFEKM RQFQLDTLCA 420
421 DTDQPVPDDV RLILDSDFER LADVQSMEEL KDVHMYFLLN YSHELPRMQA EQRRKAIQER 480
481 REAKARRQAA AAENGDDAAE AIVVPEPEDD DDPELIDYQL KQASNSSPYA VFRKAGICGF 540
541 AKHFGLTPEQ YAENLRDNYQ RNEITQESIG PTELAKQYLS PRFMTTDEVI HAAKYVVARQ 600
601 LAQEPLLRKT MREVYFDRAR INIRPTKNGM VLIDENSPVY SMKYVAKKPV SDLFGDQFIK 660
661 LMMAEEEKLL EITFLEEFEG NACANGTPGD YVEESKALYQ LDQFAKHVQE WNKLRAECVQ 720
721 LALQKWVIPD LIKELRSTLH EEAQQFVLRS CTGKLYKWLK VAPYKPQLPP DFGYEEWSTL 780
781 RGIRVLGLAY DPDHSVAAFC AVTTVEGDIS DYLRLPNILK RKNSYNLEEK AQKLADLRKL 840
841 SDFIKMKKPH IVVIGAESRD AQNIQADIKE ILHELETSEQ FPPIEVEIID NELAKIYANS 900
901 KKGESDFKEY PPLLKQAASL ARKMQDPLVE YSQLCDADDE ILCLRYHPLQ ERVPREQLLE 960
961 QLSLQFINRT SEVGLDINLM VQNSRTINLL QYICGLGPRK GQALLKLLKQ SNQRLENRTQ1020
1021 LVTVCHLGPR VFINCSGFIK IDTSSLGDST EAYVEVLDGS RVHPETYEWA RKMAIDAMEY1080
1081 DDEETNPAGA LEEILESPER LKDLDLDAFA VELERQGFGS KSITLYDIRN ELSCLYKDYR1140
1141 TPYTKPSAEE LFDMLTKETP DSFYVGKCVT AMVTGFTYRR PQGDQLDSAN PVRLDSNESW1200
1201 QCPFCHKDDF PELSEVWNHF DANACPGQPS GVRVRLENGL PGFIHIKNLS DRQVRNPEER1260
1261 VRVSQMIHVR IIKIDIDRFS VECSSRTADL KDVNNEWRPR RDNYYDYVTE EQDNRKVSDA1320
1321 KARALKRKIY ARRVIAHPSF FNKSYAEVVA MLAEADQGEV ALRPSSKSKD HLTATWKVAD1380
1381 DIFQHIDVRE EGKENDFSLG RSLWIGTEEF EDLDEIIARH IMPMALAARE LIQYKYYKPN1440
1441 MVTGDENERD VMEKLLREEK ANDPKKIHYF FTASRAMPGK FLLSYLPKTK VRHEYVTVMP1500
1501 EGYRFRGQIF DTVNSLLRWF KEHWLDPTAT PASASASNLT PLHLMRPPPT ISSSSQTSLG1560
1561 PQAPYSVTGS VTGGTPRSGI SSAVGGGGSS AYSITQSITG YGTSGSSAPG AGVSSSHYGS1620
1621 SSTPSFGAIN TPYTPSGQTP FMTPYTPHAS QTPRYGHNVP SPSSQSSSSQ RHHYGSSSGT1680
1681 GSTPRYHDMG GGGGGGVGGG GGSNAYSMQP HHQQRAKENL DWQLANDAWA RRRPQQHQSH1740
1741 QSYHAQQQHH HSQQQPHMGM SMNMGITMSL GRGTGGGGGG GYGSTPVNDY STGGGHNRGM1800
1801 SSKASVRSTP RTNASPHSMN LGDATPLYDE N |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
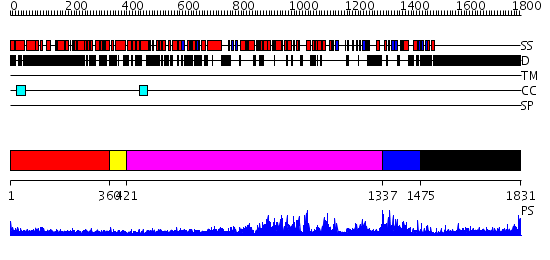
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..359] | 30.30103 | Heavy meromyosin subfragment | |
| 2 | View Details | [360..420] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [421..1336] | 1000.0 | No description for 2oceA was found. | |
| 4 | View Details | [1337..1474] | 3.91 | The Xlp protein Sap | |
| 5 | View Details | [1475..1831] | 12.221849 | No description for 1y0fB was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)