













| General Information: |
|
| Name(s) found: |
FBpp0310135
[FlyBase]
Pp2C1-PA / FBpp0070653 [FlyBase] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1427 amino acids |
Gene Ontology: |
|
| Cellular Component: |
protein serine/threonine phosphatase complex
[IEA]
|
| Biological Process: |
protein amino acid dephosphorylation
[IEA][ISS][NAS]
oxidation reduction [IEA] |
| Molecular Function: |
protein serine/threonine phosphatase activity
[IEA][ISS][NAS]
oxidoreductase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLPANNRSST STHTNTNANT INATTNTTNR CLINTAIEKT VVRLRETAAN SGTFQDSDES 60
61 SVEFDIINYY GDDGGTTPAP APATASVTRH GGSSSGNNNN NSACHPALDA SSDVVVVEPA 120
121 AVGVAQEEEE EPEQRPERIS IPIPDLAFTE MEAYAEDIVV DMEGGSPAKP LNPKKQRLNS 180
181 ATTTTINRSR GGGAAQSRLR RSAAIVPPRS IPESCASSSN SNSSSSSNSN SSSSSATGSS 240
241 ASTGNPSPCS SLGVNMRVTG QCCQGGRKYM EDQFSVAYQE SPITHELEYA FFGIYDGHGG 300
301 PEAALFAKEH LMLEIVKQKQ FWSDQDEDVL RAIREGYIAT HFAMWREQEK WPRTANGHLS 360
361 TAGTTATVAF MRREKIYIGH VGDSGIVLGY QNKGERNWRA RPLTTDHKPE SLAEKTRIQR 420
421 SGGNVAIKSG VPRVVWNRPR DPMHRGPIRR RTLVDEIPFL AVARSLGDLW SYNSRFKEFV 480
481 VSPDPDVKVV KINPSTFRCL IFGTDGLWNV VTAQEAVDSV RKEHLIGEIL NEQDVMNPSK 540
541 ALVDQALKTW AAKKMRADNT SVVTVILTPA ARNNSPTTPT RSPSAMARDN DLEVELLLEE 600
601 DDEELPTLDV ENNYPDFLIE EHEYVLDQPY SALAKRHSPP EAFRNFDYFD VDEDELDEDE 660
661 ETVEEDEEEE EEEEETKSVG ILQQSLFNPR KTWRKSTINN SWSGVTEPEP EPDPEPDRID 720
721 VLTLDMYSHT SIDKGTNYGG SIAQSSIDPA ETAENRELSE LEQHLESSYS FAESYNSLLN 780
781 EQEEQEARSR SAAAAAAAAE AAAVEAQQTT AHSASVVLDR SMLEIIQEQQ HYQQQEGYSL 840
841 TQLETRRERE RLTESWPQQP AELLELDALL QQERAEEEQV ALEQQQQREQ QMEQMEVEAI 900
901 SSSGQHEFAY PVTTATASEW CATLQEDEEE LDSTVIDIVI QPEQELQDNE VSSTLPATPT 960
961 HVEPEQIVDK MEPLKVQEML TAVEKPPSKQ EKKLPKKQET KQVAVLDTVA EMPKEDAHAV1020
1021 HYIFQRIQKV QDSEATPVAV TNSTMADALP TESSGLGGSM TAPRIRRYRN VPNENHQHMQ1080
1081 TRRRQIFKHV KPKSFIQSSA AAIVAYGDST ETVGGTAGAS GTPAAGRVGG GGGGGGGRGS1140
1141 ASGGSSPAVA ANSRRSVNVV ANASGNSASK VVPSSSSMMM TRRSHTLTAS GGVNKRQLRS1200
1201 SLCTLGLGVG VGVGLGMDLD MTKRTLRTRN VPALSGGSAT PSSNSSPASG GSSPAGFTSP1260
1261 ASPVITSRGS GSRTTASPAR RLKRSHEDRE QRMSLRRSTL SGSASGSGLV GTGGSPSNVK1320
1321 SNRLQACNGA ISARPPPSPK KLNAAVPTLA IGTRAYTAAL AAAADHLNKR WSLRSSSGNS1380
1381 GNLITAISCY SDRSRAATAA GSPGSGGGAA GPPGASLAAS TVGTRRR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
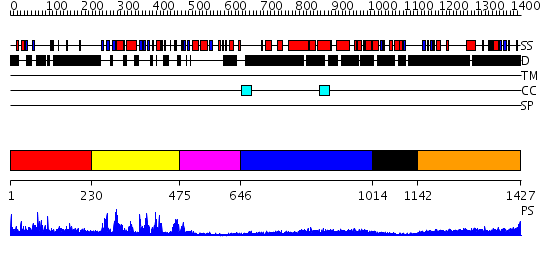
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..229] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [230..474] | 78.69897 | CRYSTAL STRUCTURE OF THE PROTEIN SERINE/THREONINE PHOSPHATASE 2C AT 2 A RESOLUTION | |
| 3 | View Details | [475..645] | 3.79 | CRYSTAL STRUCTURE OF THE PROTEIN SERINE/THREONINE PHOSPHATASE 2C AT 2 A RESOLUTION | |
| 4 | View Details | [646..1013] | 31.221849 | Heavy meromyosin subfragment | |
| 5 | View Details | [1014..1141] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1142..1427] | 6.0 | DELETION MUTANT DELTA(145-150), F151D OF CYCLODEXTRIN GLYCOSYLTRANSFERASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.64 |
Source: Reynolds et al. (2008)