













| General Information: |
|
| Name(s) found: |
FBpp0310174
[FlyBase]
mei-9-PA / FBpp0070624 [FlyBase] |
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 961 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
protein complex [IDA] |
| Biological Process: |
reciprocal meiotic recombination
[IMP]
meiotic chromosome segregation [IMP] DNA recombination [NAS][TAS] nucleotide-excision repair [IMP][ISS][NAS][TAS] resolution of meiotic recombination intermediates [IMP][TAS] female meiosis chromosome segregation [IMP] mismatch repair [IMP][NAS] DNA metabolic process [IEA] double-strand break repair [TAS] |
| Molecular Function: |
DNA binding
[IEA]
protein binding [IPI] protein heterodimerization activity [IPI] endodeoxyribonuclease activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADSCAENAA KGTENERPKE VEASADTVPQ VEEGVEEYLK RKNMVLLDYE KQMFLDLVEA 60
61 DGLLVCAKGL SYDRVVISIL KAYSDSGNLV LVINSSDWEE QYYKSKIEPK YVHEVASTAT 120
121 ERERVYLEGG LQFISTRILV VDLLKQRIPI ELISGIIVLR AHTIIESCQE AFALRLFRQK 180
181 NKTGFVKAFS SSPEAFTIGY SHVERTMRNL FVKHLYIWPR FHESVRTVLQ PWKIQSIEMH 240
241 VPISQNITSI QSHILEIMNF LVQEIKRINR TVDMEAVTVE NCVTKSFHKI LQAQLDCIWH 300
301 QLNSQTKLIV ADLKILRSLM ISTMYHDAVS AYAFMKRYRS TEYALSNSGW TLLDAAEQIF 360
361 KLSRQRVFNG QQEFEPEPCP KWQTLTDLLT KEIPGDMRRS RRSEQQPKVL ILCQDARTCH 420
421 QLKQYLTQGG PRFLLQQALQ HEVPVGKLSD NYAKESQTRS APPKNVSSNK ELRREEVSGS 480
481 QPPLAGMDEL AQLLSESETE GQHFEESYML TMTQPVEVGP AAIDIKPDPD VSIFETIPEL 540
541 EQFDVTAALA SVPHQPYICL QTFKTEREGS MALEHMLEQL QPHYVVMYNM NVTAIRQLEV 600
601 FEARRRLPPA DRMKVYFLIH ARTVEEQAYL TSLRREKAAF EFIIDTKSKM VIPKYQDGKT 660
661 DEAFLLLKTY DDEPTDENAK SRQAGGQAPQ ATKETPKVIV DMREFRSDLP CLIHKRGLEV 720
721 LPLTITIGDY ILTPDICVER KSISDLIGSL NSGRLYNQCV QMQRHYAKPI LLIEFDQNKP 780
781 FHLQGKFMLS QQTSMANADI VQKLQLLTLH FPKLRLIWSP SPYATAQLFE ELKLGKPEPD 840
841 PQTAAALGSD EPTAGEQLHF NSGIYDFLLR LPGVHTRNIH GLLRKGGSLR QLLLRSQKEL 900
901 EELLQSQESA KLLYDILHVA HLPEKDEVTG STALLAASKQ FGAGSHNRFR MAAAASRRGR 960
961 R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
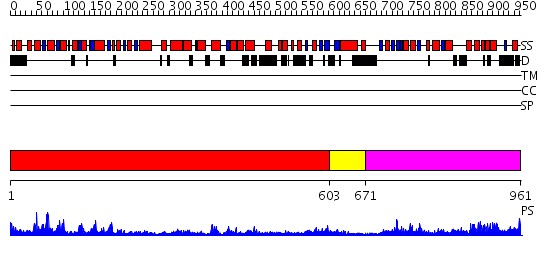
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..602] | 64.522879 | Crystal structure of Pyrococcus furiosus Hef helicase domain | |
| 2 | View Details | [603..670] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [671..961] | 28.0 | XPF from Aeropyrum pernix, complex with DNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)